3QEE
 
 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | ACETATE ION, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
3QED
 
 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, CALCIUM ION, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
3QEF
 
 | | The structure and function of an arabinan-specific alpha-1,2-arabinofuranosidase identified from screening the activities of bacterial GH43 glycoside hydrolases | | Descriptor: | 1,2-ETHANEDIOL, Beta-xylosidase/alpha-L-arabinfuranosidase, gly43N, ... | | Authors: | Cartmell, A, Mckee, L.S, Pena, M, Larsbrink, J, Brumer, H, Lewis, R.J, Viks-Nielsen, A, Gilbert, H.J, Marles-Wright, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-02-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | The Structure and Function of an Arabinan-specific {alpha}-1,2-Arabinofuranosidase Identified from Screening the Activities of Bacterial GH43 Glycoside Hydrolases.
J.Biol.Chem., 286, 2011
|
|
2CDG
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5B) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
2CDE
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide specific T cell receptors - iNKT-TCR | | Descriptor: | INKT-TCR | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
2CDF
 
 | | Structure and binding kinetics of three different human CD1d-alpha- Galactosylceramide-specific T cell receptors (TCR 5E) | | Descriptor: | TCR 5E | | Authors: | Gadola, S.D, Koch, M, Marles-Wright, J, Lissin, N.M, Sheperd, D, Matulis, G, Harlos, K, Villiger, P.M, Stuart, D.I, Jakobsen, B.K, Cerundolo, V, Jones, E.Y. | | Deposit date: | 2006-01-23 | | Release date: | 2006-03-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structrue and Binding Kinetics of Three Different Human Cd1D-Alpha-Galactosylceramide-Specific T Cell Receptors
J.Exp.Med., 203, 2006
|
|
5DBV
 
 | | Structure of a C269A mutant of propionaldehyde dehydrogenase from the Clostridium phytofermentans fucose utilisation bacterial microcompartment | | Descriptor: | ACETATE ION, Aldehyde Dehydrogenase, COENZYME A, ... | | Authors: | Tuck, L.R, Altenbach, K, Ang, T.F, Crawshaw, A.D, Campopiano, D.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2015-08-22 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Insight into Coenzyme A cofactor binding and the mechanism of acyl-transfer in an acylating aldehyde dehydrogenase from Clostridium phytofermentans.
Sci Rep, 6, 2016
|
|
5DA5
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 | | Descriptor: | CALCIUM ION, FE (III) ION, GLYCOLIC ACID, ... | | Authors: | He, D, Vanden Hehier, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2015-08-19 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
7NWR
 
 | | Structure of BT1526, a myo-inositol-1-phosphate synthase | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Basle, A, Tang, G, Marles-Wright, J, Campopiano, D. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of inositol lipid metabolism in gut-associated Bacteroidetes.
Nat Microbiol, 7, 2022
|
|
2VSI
 
 | | Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, 4-AMINO-1-{5-O-[(R)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]-ALPHA-D-ARABINOFURANOSYL}PYRIMIDIN-2(1H)-ONE, CALCIUM ION | | Authors: | Baur, S, Marles-Wright, J, Buckenmaier, S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Synthesis of Cdp-Activated Ribitol for Teichoic Acid Precursors in Streptococcus Pneumoniae.
J.Bacteriol., 191, 2009
|
|
2VSH
 
 | | Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae | | Descriptor: | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baur, S, Marles-Wright, J, Buckenmaier, S, Lewis, R.J, Vollmer, W. | | Deposit date: | 2008-04-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis of Cdp-Activated Ribitol for Teichoic Acid Precursors in Streptococcus Pneumoniae.
J.Bacteriol., 191, 2009
|
|
6G1F
 
 | |
6HBS
 
 | |
6HBV
 
 | |
5L8B
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E62A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5L89
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A | | Descriptor: | CALCIUM ION, Rru_A0973 | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5L8G
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant H65A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.974 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
5L8F
 
 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A, E62A, H65A. | | Descriptor: | MAGNESIUM ION, Rru_A0973, TRIETHYLENE GLYCOL | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E32A, E62A, H65A.
To be published
|
|
4AXJ
 
 | | Structure of the Clostridium difficile EutM protein | | Descriptor: | ETHANOLAMINE CARBOXYSOME STRUCTURAL PROTEIN, SULFATE ION | | Authors: | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2012-06-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
4AXI
 
 | | Structure of the Clostridium difficile EutS protein | | Descriptor: | ETHANOLAMINE CARBOXYSOME STRUCTURAL PROTEIN, GLYCEROL | | Authors: | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2012-06-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
4AXO
 
 | | Structure of the Clostridium difficile EutQ protein | | Descriptor: | ETHANOLAMINE UTILIZATION PROTEIN, MAGNESIUM ION | | Authors: | Pitts, A.C, Tuck, L.R, Faulds-Pain, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2012-06-13 | | Release date: | 2012-06-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Insight Into the Clostridium Difficile Ethanolamine Utilisation Microcompartment.
Plos One, 7, 2012
|
|
3TFL
 
 | | LytR-Cps2a-Psr family protein with bound octaprenyl pyrophosphate lipid | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, CPS2A, ... | | Authors: | Kawai, Y, Marles-Wright, J, Cleverley, R.M, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Khai Bui, N, Hoyland, C.N, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2011-08-16 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A widespread family of bacterial cell wall assembly proteins.
Embo J., 30, 2011
|
|
3TEP
 
 | | LytR-CPS2a-Psr family protein with bound octaprenyl pyrophosphate lipid and magnesium ion | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, CPS2A, ... | | Authors: | Kawai, Y, Marles-Wright, J, Cleverley, R.M, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Khai Bui, N, Hoyland, C.N, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | A widespread family of bacterial cell wall assembly proteins.
Embo J., 30, 2011
|
|
3TEL
 
 | | LytR-CPS2A-Psr family protein with bound octaprenyl pyrophosphate lipid and manganese ion | | Descriptor: | (2Z,6Z,10Z,14Z,18Z,22Z,26Z)-3,7,11,15,19,23,27,31-octamethyldotriaconta-2,6,10,14,18,22,26,30-octaen-1-yl trihydrogen diphosphate, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kawai, Y, Marles-Wright, J, Cleverley, R.M, Emmins, R, Ishikawa, S, Kuwano, M, Heinz, N, Khai Bui, N, Hoyland, C.N, Ogasawara, N, Lewis, R.J, Vollmer, W, Daniel, R.A, Errington, J. | | Deposit date: | 2011-08-15 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A widespread family of bacterial cell wall assembly proteins.
Embo J., 30, 2011
|
|
3ZT9
 
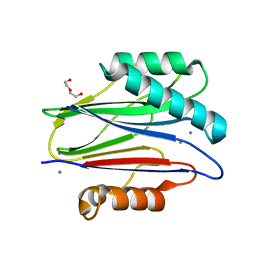 | | The bacterial stressosome: a modular system that has been adapted to control secondary messenger signaling | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, SERINE PHOSPHATASE | | Authors: | Quin, M.B, Berrisford, J.M, Newman, J.A, Basle, A, Lewis, R.J, Marles-Wright, J. | | Deposit date: | 2011-07-06 | | Release date: | 2012-02-22 | | Last modified: | 2018-10-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Bacterial Stressosome: A Modular System that Has Been Adapted to Control Secondary Messenger Signaling.
Structure, 20, 2012
|
|
