4D1B
 
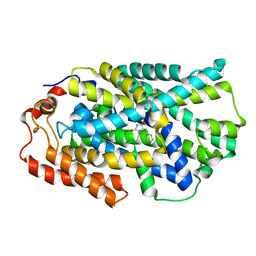 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH BENZYL-HYDANTOIN | | Descriptor: | (5S)-5-benzylimidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Brueckner, F, Geng, T, Weyand, S, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
4D1A
 
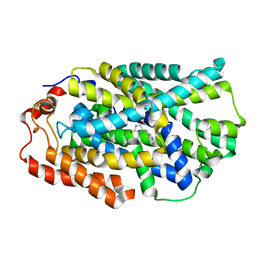 | | STRUCTURE OF MHP1, A NUCLEOBASE-CATION-SYMPORT-1 FAMILY TRANSPORTER, IN A CLOSED CONFORMATION WITH INDOLYLMETHYL-HYDANTOIN | | Descriptor: | (5S)-5-(1H-indol-3-ylmethyl)imidazolidine-2,4-dione, HYDANTOIN TRANSPORT PROTEIN, SODIUM ION | | Authors: | Weyand, S, Brueckner, F, Geng, T, Drew, D, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Molecular Mechanism of Ligand Recognition by Membrane Transport Protein, Mhp1.
Embo J., 33, 2014
|
|
5I6C
 
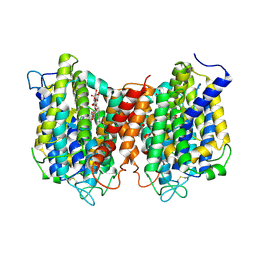 | | The structure of the eukaryotic purine/H+ symporter, UapA, in complex with Xanthine | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Uric acid-xanthine permease, XANTHINE | | Authors: | Alguel, Y, Amillis, S, Leung, J, Lambrinidis, G, Capaldi, S, Scull, N.J, Craven, G, Iwata, S, Armstrong, A, Mikros, E, Diallinas, G, Cameron, A.D, Byrne, B. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of eukaryotic purine/H(+) symporter UapA suggests a role for homodimerization in transport activity.
Nat Commun, 7, 2016
|
|
7APE
 
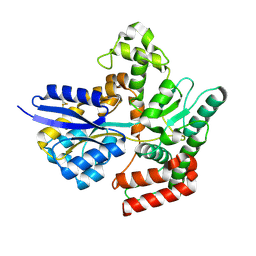 | | Crystal structure of LpqY from Mycobacterium thermoresistible in complex with trehalose | | Descriptor: | Lipoprotein (Sugar-binding) lpqY, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Furze, C.M, Guy, C.M, Angula, J, Cameron, A.D, Fullam, E. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of trehalose recognition by the mycobacterial LpqY-SugABC transporter.
J.Biol.Chem., 296, 2021
|
|
6FV3
 
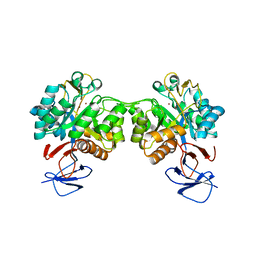 | | Crystal structure of N-acetyl-D-glucosamine-6-phosphate deacetylase from Mycobacterium smegmatis. | | Descriptor: | N-acetylglucosamine-6-phosphate deacetylase, ZINC ION | | Authors: | Ahangar, M.S, Furze, C.M, Guy, C.S, Cooper, C, Maskew, K.S, Graham, B, Cameron, A.D, Fullam, E. | | Deposit date: | 2018-02-28 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and functional determination of homologs of theMycobacterium tuberculosis N-acetylglucosamine-6-phosphate deacetylase (NagA).
J. Biol. Chem., 293, 2018
|
|
6FV4
 
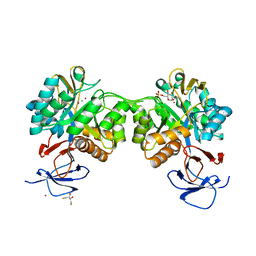 | | The structure of N-acetyl-D-glucosamine-6-phosphate deacetylase D267A mutant from Mycobacterium smegmatis in complex with N-acetyl-D-glucosamine-6-phosphate | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, CADMIUM ION, ... | | Authors: | Ahangar, M.S, Furze, C.M, Guy, C.S, Cooper, C, Maskew, K.S, Graham, B, Cameron, A.D, Fullam, E. | | Deposit date: | 2018-03-01 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural and functional determination of homologs of theMycobacterium tuberculosis N-acetylglucosamine-6-phosphate deacetylase (NagA).
J. Biol. Chem., 293, 2018
|
|
2V7A
 
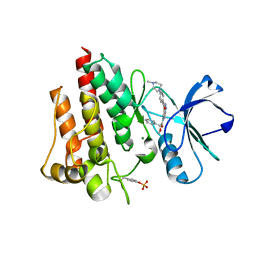 | | Crystal structure of the T315I Abl mutant in complex with the inhibitor PHA-739358 | | Descriptor: | MAGNESIUM ION, N-[(3E)-5-[(2R)-2-METHOXY-2-PHENYLACETYL]PYRROLO[3,4-C]PYRAZOL-3(5H)-YLIDENE]-4-(4-METHYLPIPERAZIN-1-YL)BENZAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL1 | | Authors: | Modugno, M, Casale, E, Soncini, C, Rosettani, P, Colombo, R, Lupi, R, Rusconi, L, Fancelli, D, Carpinelli, P, Cameron, A.D, Isacchi, A, Moll, J. | | Deposit date: | 2007-07-27 | | Release date: | 2007-09-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the T315I Abl Mutant in Complex with the Aurora Kinases Inhibitor Pha-739358.
Cancer Res., 67, 2007
|
|
3VG9
 
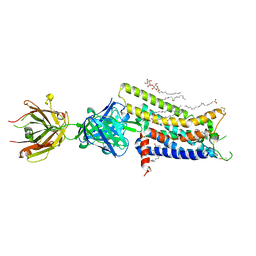 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 2.7 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
6SCT
 
 | | Cryo-EM structure of the consensus triskelion hub of the clathrin coat complex | | Descriptor: | Clathrin heavy chain, Clathrin light chain | | Authors: | Morris, K.L, Cameron, A.D, Sessions, R, Smith, C.J. | | Deposit date: | 2019-07-25 | | Release date: | 2019-10-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Cryo-EM of multiple cage architectures reveals a universal mode of clathrin self-assembly.
Nat.Struct.Mol.Biol., 26, 2019
|
|
3VGA
 
 | | Crystal structure of human adenosine A2A receptor with an allosteric inverse-agonist antibody at 3.1 A resolution | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, antibody fab fragment heavy chain, ... | | Authors: | Hino, T, Arakawa, T, Iwanari, H, Yurugi-Kobayashi, T, Ikeda-Suno, C, Nakada-Nakura, Y, Kusano-Arai, O, Weyand, S, Shimamura, T, Nomura, N, Cameron, A.D, Kobayashi, T, Hamakubo, T, Iwata, S, Murata, T. | | Deposit date: | 2011-08-04 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody
Nature, 482, 2012
|
|
6R1B
 
 | | Crystal structure of UgpB from Mycobacterium tuberculosis in complex with glycerophosphocholine | | Descriptor: | 2-(((R)-2,3-DIHYDROXYPROPYL)PHOSPHORYLOXY)-N,N,N-TRIMETHYLETHANAMINIUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fenn, J, Nepravishta, R, Guy, C.S, Harrison, J, Angulo, J, Cameron, A.D, Fullam, E. | | Deposit date: | 2019-03-14 | | Release date: | 2019-09-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27000213 Å) | | Cite: | Structural Basis of Glycerophosphodiester Recognition by theMycobacterium tuberculosisSubstrate-Binding Protein UgpB.
Acs Chem.Biol., 14, 2019
|
|
2JLN
 
 | | Structure of Mhp1, a nucleobase-cation-symport-1 family transporter | | Descriptor: | MERCURY (II) ION, MHP1, SODIUM ION | | Authors: | Weyand, S, Shimamura, T, Yajima, S, Suzuki, S, Mirza, O, Krusong, K, Carpenter, E.P, Rutherford, N.G, Hadden, J.M, O'Reilly, J, Ma, P, Saidijam, M, Patching, S.G, Hope, R.J, Norbertczak, H.T, Roach, P.C.J, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2008-09-11 | | Release date: | 2008-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure and Molecular Mechanism of a Nucleobase-Cation-Symport-1 Family Transporter.
Science, 322, 2008
|
|
4GAV
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with quinone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase, UBIQUINONE-2 | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G9K
 
 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-24 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4GAP
 
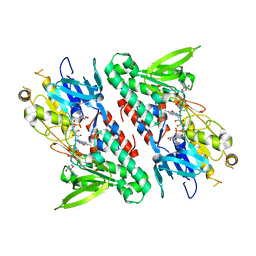 | | Structure of the Ndi1 protein from Saccharomyces cerevisiae in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Rotenone-insensitive NADH-ubiquinone oxidoreductase | | Authors: | Iwata, M, Lee, Y, Yamashita, T, Yagi, T, Iwata, S, Cameron, A.D, Maher, M.J. | | Deposit date: | 2012-07-25 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of the yeast NADH dehydrogenase (Ndi1) reveals overlapping binding sites for water- and lipid-soluble substrates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4YZF
 
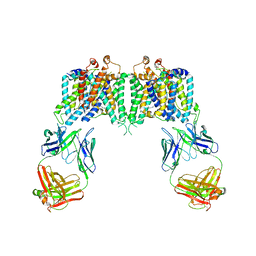 | | Crystal structure of the anion exchanger domain of human erythrocyte Band 3 | | Descriptor: | 2,2'-ethane-1,2-diylbis{5-[(sulfanylmethyl)amino]benzenesulfonic acid}, Band 3 anion transport protein, FAB fragment of Immunoglobulin (IgG) molecule | | Authors: | Alguel, Y, Arakawa, T, Yugiri, T.K, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Suno, C.I, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A.D, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-03-25 | | Release date: | 2015-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the anion exchanger domain of human erythrocyte band 3.
Science, 350, 2015
|
|
1H8V
 
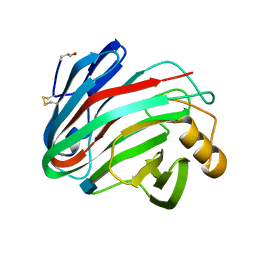 | | The X-ray Crystal Structure of the Trichoderma reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Shaw, A, Ropp, T.H, Wu, S, Bott, R, Cameron, A.D, Stahlberg, J, Mitchinson, C, Jones, T.A. | | Deposit date: | 2001-02-16 | | Release date: | 2001-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The X-Ray Crystal Structure of the Trichoderma Reesei Family 12 Endoglucanase 3, Cel12A, at 1.9 A Resolution
J.Mol.Biol., 308, 2001
|
|
2XRU
 
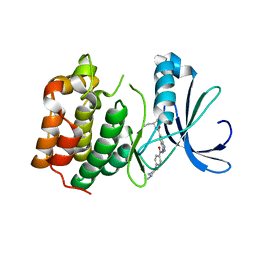 | | AURORA-A T288E COMPLEXED WITH PHA-828300 | | Descriptor: | 3-({[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]CARBONYL}AMINO)-N-[(1R)-1-PHENYLPROPYL]-1H-THIENO[3,2-C]PYRAZOLE-5-CARBOXAMIDE, SERINE/THREONINE-PROTEIN KINASE 6 | | Authors: | Bindi, S, Fancelli, D, Alli, C, Berta, D, Bertrand, J.A, Cameron, A.D, Cappella, P, Carpinelli, P, Cervi, G, Croci, W, D'Anello, M, Forte, B, LauraGiorgini, M, Marsiglio, A, Moll, J, Pesenti, E, Pittala, V, Pulici, M, Riccardi-Sirtori, F, Roletto, F, Soncini, C, Storici, P, Varasi, M, Volpi, D, Zugnoni, P, Vianello, P. | | Deposit date: | 2010-09-22 | | Release date: | 2010-09-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Thieno[3,2-C]Pyrazoles: A Novel Class of Aurora Inhibitors with Favorable Antitumor Activity.
Bioorg.Med.Chem., 18, 2010
|
|
2XVM
 
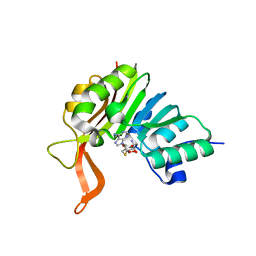 | | Crystal structure of the tellurite detoxification protein TehB from E. coli in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, TELLURITE RESISTANCE PROTEIN TEHB | | Authors: | Choudhury, H.G, Cameron, A.D, Iwata, S, Beis, K. | | Deposit date: | 2010-10-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure and Mechanism of the Chalcogen Detoxifying Protein Tehb from Escherichia Coli.
Biochem.J., 435, 2011
|
|
2XUT
 
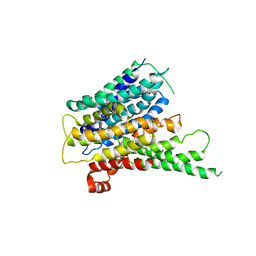 | | Crystal structure of a proton dependent oligopeptide (POT) family transporter. | | Descriptor: | PROTON/PEPTIDE SYMPORTER FAMILY PROTEIN | | Authors: | Newstead, S, Drew, D, Cameron, A.D, Postis, V.L, Xia, X, Fowler, P.W, Ingram, J.C, Carpenter, E.P, Sansom, M.S.P, McPherson, M.J, Baldwin, S.A, Iwata, S. | | Deposit date: | 2010-10-21 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal Structure of a Prokaryotic Homologue of the Mammalian Oligopeptide-Proton Symporters, Pept1 and Pept2.
Embo J., 30, 2011
|
|
2WIH
 
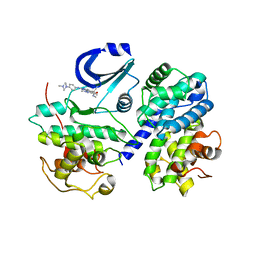 | | STRUCTURE OF CDK2-CYCLIN A WITH PHA-848125 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN-A2, N,1,4,4-TETRAMETHYL-8-{[4-(4-METHYLPIPERAZIN-1-YL)PHENYL]AMINO}-4,5-DIHYDRO-1H-PYRAZOLO[4,3-H]QUINAZOLINE-3-CARBOXAMIDE, ... | | Authors: | Brasca, M.G, Amboldi, N, Ballinari, D, Cameron, A.D, Casale, E, Cervi, G, Colombo, M, Colotta, F, Croci, V, Dalessio, R, Fiorentini, F, Isacchi, A, Mercurio, C, Moretti, W, Panzeri, A, Pastori, W, Pevarello, P, Quartieri, F, Roletto, F, Traquandi, G, Vianello, P, Vulpetti, A, Ciomei, M. | | Deposit date: | 2009-05-13 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of N,1,4,4-Tetramethyl-8-{[4-(4-Methylpiperazin-1-Yl)Phenyl]Amino}-4,5-Dihydro-1H-Pyrazolo[4,3-H]Quinazoline-3-Carboxamide (Pha-848125), a Potent, Orally Available Cyclin Dependent Kinase Inhibitor.
J.Med.Chem., 52, 2009
|
|
2X79
 
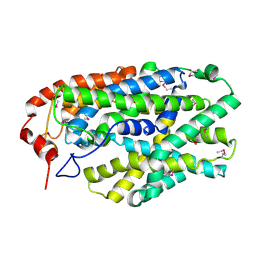 | | Inward facing conformation of Mhp1 | | Descriptor: | Hydantoin permease | | Authors: | Shimamura, T, Weyand, S, Beckstein, O, Rutherford, N.G, Hadden, J.M, Sharples, D, Sansom, M.S.P, Iwata, S, Henderson, P.J.F, Cameron, A.D. | | Deposit date: | 2010-02-25 | | Release date: | 2010-05-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Molecular Basis of Alternating Access Membrane Transport by the Sodium-Hydantoin Transporter Mhp1.
Science, 328, 2010
|
|
2XVA
 
 | | Crystal structure of the tellurite detoxification protein TehB from E. coli in complex with sinefungin | | Descriptor: | SINEFUNGIN, TELLURITE RESISTANCE PROTEIN TEHB, ZINC ION | | Authors: | Choudhury, H.G, Cameron, A.D, Iwata, S, Beis, K. | | Deposit date: | 2010-10-25 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Mechanism of the Chalcogen Detoxifying Protein Tehb from Escherichia Coli.
Biochem.J., 435, 2011
|
|
4APS
 
 | | Crystal structure of a POT family peptide transporter in an inward open conformation. | | Descriptor: | CADMIUM ION, DI-OR TRIPEPTIDE H+ SYMPORTER | | Authors: | Solcan, N, Kwok, J, Fowler, P.W, Cameron, A.D, Drew, D, Iwata, S, Newstead, S. | | Deposit date: | 2012-04-05 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Alternating Access Mechanism in the Pot Family of Oligopeptide Transporters.
Embo J., 31, 2012
|
|
7QET
 
 | | human Connexin 26 dodecamer at 20mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
