8T6J
 
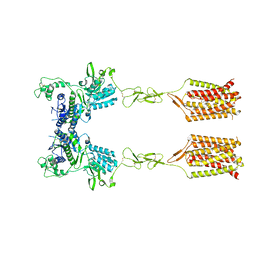 | | CDPPB-bound inactive mGlu5 | | Descriptor: | 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-16 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8TAO
 
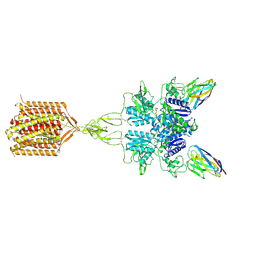 | | Quis and CDPPB bound active mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5, ... | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-27 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T7H
 
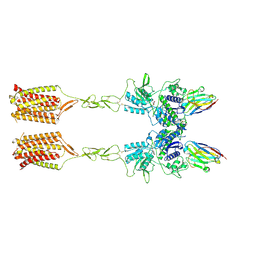 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nanobody 43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-20 | | Release date: | 2023-10-11 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
4F7P
 
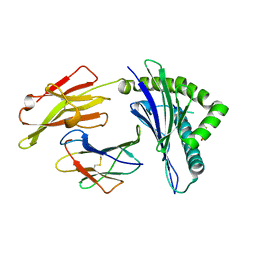 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009H1N1 PB1 (496-505) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, Gao, G.F. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
3EQE
 
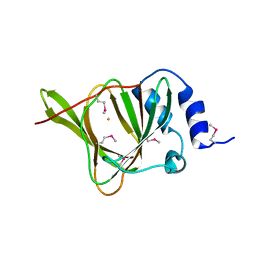 | | Crystal structure of the YubC protein from Bacillus subtilis. Northeast Structural Genomics Consortium target SR112. | | Descriptor: | FE (III) ION, Putative cystein dioxygenase | | Authors: | Vorobiev, S.M, Su, M, Seetharaman, J, Benach, J, Forouhar, F, Clayton, G, Cooper, B, Wang, H, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-09-30 | | Release date: | 2008-10-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Crystal structure of the YubC protein from Bacillus subtilis.
To be Published
|
|
4F7M
 
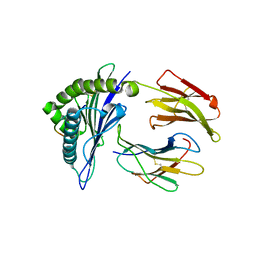 | | Crystal Structure of HLA-A*2402 Complexed with a Newly Identified Peptide from 2009 H1N1 PA (649-658) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-24 alpha chain, ... | | Authors: | Liu, J, Zhang, S, Tan, S, Yi, Y, Wu, B, Zhu, F, Wang, H, Qi, J, George, F.G. | | Deposit date: | 2012-05-16 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cross-Allele Cytotoxic T Lymphocyte Responses against 2009 Pandemic H1N1 Influenza A Virus among HLA-A24 and HLA-A3 Supertype-Positive Individuals.
J.Virol., 86, 2012
|
|
3F08
 
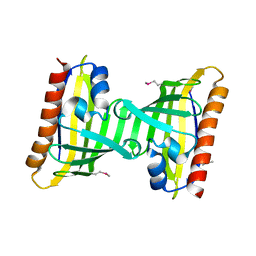 | | Crystal structure of the putative uncharacterized protein Q6HG14 from Bacilllus thuringiensis. Northeast Structural Genomics Consortium target BuR153. | | Descriptor: | uncharacterized protein Q6HG14 | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Wang, H, Mao, L, Ciccosanti, C, Xiao, R, Nair, R, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the putative uncharacterized protein Q6HG14 from Bacilllus thuringiensis. Northeast Structural Genomics Consortium target BuR153.
To be Published
|
|
4C5Z
 
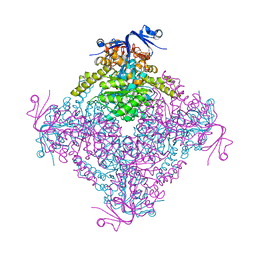 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4C60
 
 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4C5Y
 
 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE, ZINC ION | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4C65
 
 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4KUL
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain V83P mutant | | Descriptor: | Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUI
 
 | | Crystal structure of N-terminal acetylated yeast Sir3 BAH domain | | Descriptor: | ACETIC ACID, ISOPROPYL ALCOHOL, Regulatory protein SIR3 | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KUD
 
 | | Crystal structure of N-terminal acetylated Sir3 BAH domain D205N mutant in complex with yeast nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.1, Histone H3, ... | | Authors: | Yang, D, Fang, Q, Wang, M, Ren, R, Wang, H, He, M, Sun, Y, Yang, N, Xu, R.M. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | N alpha-acetylated Sir3 stabilizes the conformation of a nucleosome-binding loop in the BAH domain.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3KOJ
 
 | | Crystal structure of the SSB domain of Q5N255_SYNP6 protein from Synechococcus sp. Northeast Structural Genomics Consortium Target SnR59a. | | Descriptor: | uncharacterized protein ycf41 | | Authors: | Vorobiev, S, Su, M, Seetharaman, J, Wang, H, Foote, L.E, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-11-13 | | Release date: | 2009-11-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Crystal structure of the SSB domain of Q5N255_SYNP6 protein from Synechococcus sp.
To be Published
|
|
3LML
 
 | | Crystal Structure of the sheath tail protein Lin1278 from Listeria innocua, Northeast Structural Genomics Consortium Target LkR115 | | Descriptor: | Lin1278 protein | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Mao, M, Xiao, R, Patel, D.J, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-31 | | Release date: | 2010-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Northeast Structural Genomics Consortium Target LkR115
To be Published
|
|
1A8O
 
 | | HIV CAPSID C-TERMINAL DOMAIN | | Descriptor: | HIV CAPSID | | Authors: | Gamble, T.R, Yoo, S, Vajdos, F.F, Von Schwedler, U.K, Worthylake, D.K, Wang, H, Mccutcheon, J.P, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-03-27 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein.
Science, 278, 1997
|
|
1A43
 
 | | STRUCTURE OF THE HIV-1 CAPSID PROTEIN DIMERIZATION DOMAIN AT 2.6A RESOLUTION | | Descriptor: | HIV-1 CAPSID | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-02-10 | | Release date: | 1999-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BAJ
 
 | | HIV-1 CAPSID PROTEIN C-TERMINAL FRAGMENT PLUS GAG P2 DOMAIN | | Descriptor: | GAG POLYPROTEIN | | Authors: | Worthylake, D.K, Wang, H, Yoo, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 1998-04-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
3L46
 
 | | Crystal structure of the second BRCT domain of epithelial cell transforming 2 (ECT2) | | Descriptor: | Protein ECT2, UNKNOWN ATOM OR ION | | Authors: | Nedyalkova, L, Tempel, W, Tong, Y, Crombet, L, Wang, H, Guan, X, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | Crystal structure of the second BRCT domain of epithelial cell transforming 2 (ECT2)
to be published
|
|
1AUM
 
 | | HIV CAPSID C-TERMINAL DOMAIN (CAC146) | | Descriptor: | HIV CAPSID | | Authors: | Hill, C.P, Gamble, T.R, Yoo, S, Vajdos, F.F, Von Schwedler, U.K, Worthylake, D.K, Wang, H, Mccutcheon, J.P, Sundquist, W.I. | | Deposit date: | 1997-08-29 | | Release date: | 1998-01-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein.
Science, 278, 1997
|
|
3KH0
 
 | | Crystal structure of the Ras-association (RA) domain of RALGDS | | Descriptor: | Ral guanine nucleotide dissociation stimulator, UNKNOWN ATOM OR ION | | Authors: | Shen, Y, Tempel, W, Wang, H, Tong, Y, Guan, X, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-29 | | Release date: | 2009-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the Ras-association (RA) domain of RALGDS
to be published
|
|
9K9U
 
 | | MPXV DNA polymerase complex in editing state 1 | | Descriptor: | DNA (25-MER), DNA (38-MER), DNA polymerase, ... | | Authors: | Xie, Y.F, Kuai, L, Peng, Q, Wang, Q, Wang, H, Li, X.M, Qi, J.X, Ding, Q, Shi, Y, Gao, F. | | Deposit date: | 2024-10-27 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | MPXV DNA polymerase complex in editing state 1
To Be Published
|
|
3MA5
 
 | | Crystal structure of the tetratricopeptide repeat domain protein Q2S6C5_SALRD from Salinibacter ruber. Northeast Structural Genomics Consortium Target SrR115c. | | Descriptor: | Tetratricopeptide repeat domain protein | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Wang, H, Foote, E.L, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-23 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the tetratricopeptide repeat domain protein Q2S6C5_SALRD from Salinibacter ruber.
To be Published
|
|
1BC9
 
 | | CYTOHESIN-1/B2-1 SEC7 DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOHESIN-1 | | Authors: | Betz, S.F, Schnuchel, A, Wang, H, Olejniczak, E.T, Meadows, R.P, Fesik, S.W. | | Deposit date: | 1998-05-06 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cytohesin-1 (B2-1) Sec7 domain and its interaction with the GTPase ADP ribosylation factor 1.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
