1VH4
 
 | |
1VHO
 
 | |
1VHY
 
 | |
1VIY
 
 | | Crystal structure of dephospho-CoA kinase | | Descriptor: | Dephospho-CoA kinase, SULFATE ION | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural analysis of a set of proteins resulting from a bacterial genomics project
Proteins, 60, 2005
|
|
1VHA
 
 | |
1J6V
 
 | | CRYSTAL STRUCTURE OF D. RADIODURANS LUXS, C2 | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
4KDS
 
 | |
2H9X
 
 | | NMR structure for the CgNa toxin from the sea anemone Condylactis gigantea | | Descriptor: | Toxin CgNa | | Authors: | Lopez-Mendez, B, Perez-Castells, J, Gimenez-Gallego, G, Jimenez-Barbero, J. | | Deposit date: | 2006-06-12 | | Release date: | 2007-06-05 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | CgNa, a type I toxin from the giant Caribbean sea anemone Condylactis gigantea shows structural similarities to both type I and II toxins, as well as distinctive structural and functional properties(1).
Biochem.J., 406, 2007
|
|
3D30
 
 | | Structure of an expansin like protein from Bacillus Subtilis at 1.9A resolution | | Descriptor: | Expansin like protein, FORMIC ACID, GLYCEROL | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and activity of Bacillus subtilis YoaJ (EXLX1), a bacterial expansin that promotes root colonization.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3GLZ
 
 | | Human Transthyretin (TTR) complexed with(E)-3-(2-(trifluoromethyl)benzylideneaminooxy)propanoic acid (inhibitor 11) | | Descriptor: | 3-[({(1E)-[2-(trifluoromethyl)phenyl]methylidene}amino)oxy]propanoic acid, Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Orlandini, E, Sacchettini, J.C. | | Deposit date: | 2009-03-12 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Novel transthyretin amyloid fibril formation inhibitors: synthesis, biological evaluation, and X-ray structural analysis
Plos One, 4, 2009
|
|
6P2D
 
 | | Structure of mouse ketohexokinase-C in complex with fructose and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ketohexokinase, NITRATE ION, ... | | Authors: | Gasper, W.C, Allen, K.N, Tolan, D.R. | | Deposit date: | 2019-05-21 | | Release date: | 2020-06-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Michaelis-like complex of mouse ketohexokinase isoform C
ACTA CRYSTALLOGR.,SECT.D, 2024
|
|
3I25
 
 | | Potent Beta-Secretase 1 hydroxyethylene Inhibitor | | Descriptor: | Beta-secretase 1, N-[(2S,3S,5R)-1-(3,5-difluorophenoxy)-3-hydroxy-5-(2-methoxyethoxy)-6-[[(2S)-3-methyl-1-oxo-1-(phenylmethylamino)butan-2-yl]amino]-6-oxo-hexan-2-yl]-5-(methyl-methylsulfonyl-amino)-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Lindberg, J.D, Borkakoti, N, Nystrom, S. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of potent BACE-1 inhibitors containing a new hydroxyethylene (HE) scaffold: exploration of P1' alkoxy residues and an aminoethylene (AE) central core
Bioorg.Med.Chem., 18, 2010
|
|
3ICH
 
 | |
6YBK
 
 | | Structure of MBP-Mcl-1 in complex with compound 4d | | Descriptor: | (2~{R})-2-[5-[3-chloranyl-2-methyl-4-[2-(4-methylpiperazin-1-yl)ethoxy]phenyl]-6-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4-yl]oxy-3-[2-(pyrazin-2-ylmethoxy)phenyl]propanoic acid, CHLORIDE ION, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, ... | | Authors: | Dokurno, P, Surgenor, A.E, Murray, J.B. | | Deposit date: | 2020-03-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of S64315, a Potent and Selective Mcl-1 Inhibitor.
J.Med.Chem., 63, 2020
|
|
8QW7
 
 | | Crystal Structure of compound 4 in complex with KRAS G12V C118S GDP and pVHL:ElonginC:ElonginB | | Descriptor: | (2S,4R)-1-[(2R)-2-[3-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2-oxazol-5-yl]-3-methyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
8QVU
 
 | | Crystal Structure of ligand ACBI3 in complex with KRAS G12D C118S GDP and pVHL:ElonginC:ElonginB complex | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Wijaya, A.J, Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
8QU8
 
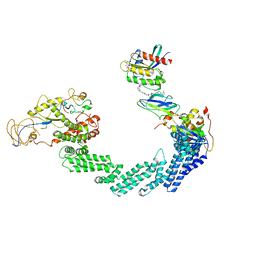 | | PROTAC-mediated complex of KRAS with VHL/Elongin-B/Elongin-C/Cullin-2/Rbx1 | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Fischer, G, Peter, D, Arce-Solano, S. | | Deposit date: | 2023-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
8QW6
 
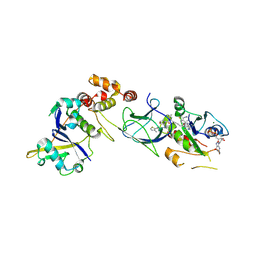 | | Crystal Structure of compound 3 in complex with KRAS G12V C118S GDP and pVHL:ElonginC:ElonginB | | Descriptor: | (2S,4R)-1-[(2S)-2-[6-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]hexanoylamino]-3,3-dimethyl-butanoyl]-N-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Zollman, D, Farnaby, W, Ciulli, A. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
8QUG
 
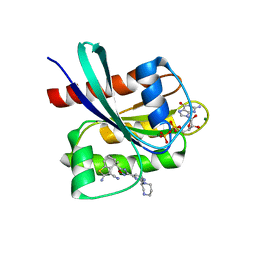 | | KRAS-G12C in Complex with Compound 1 | | Descriptor: | (4S)-2-azanyl-4-methyl-4-[3-[2-[(2S)-2-methyl-1,4-diazepan-1-yl]pyrimidin-4-yl]-1,2,4-oxadiazol-5-yl]-6,7-dihydro-5H-1-benzothiophene-3-carbonitrile, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fischer, G, Kratochvil, B. | | Deposit date: | 2023-10-16 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
8EFN
 
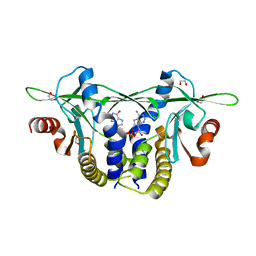 | | Structure of Sp-STING3 from Stylophora pistillata coral in complex with 3',3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon genes protein | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8EFM
 
 | | Structure of coral STING receptor from Stylophora pistillata in complex with 2',3'-cGAMP | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Li, Y, Slavik, K.M, Morehouse, B.R, Mears, K, Kranzusch, P.J. | | Deposit date: | 2022-09-08 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
6HF2
 
 | | The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycosyl hydrolase family 26 | | Authors: | Bagenholm, V, Logan, D.T, Stalbrand, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A surface-exposed GH26 beta-mannanase fromBacteroides ovatus: Structure, role, and phylogenetic analysis ofBoMan26B.
J.Biol.Chem., 294, 2019
|
|
6HF4
 
 | | The structure of BoMan26B, a GH26 beta-mannanase from Bacteroides ovatus, complexed with G1M4 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Glycosyl hydrolase family 26, ... | | Authors: | Bagenholm, V, Logan, D.T, Stalbrand, H. | | Deposit date: | 2018-08-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A surface-exposed GH26 beta-mannanase fromBacteroides ovatus: Structure, role, and phylogenetic analysis ofBoMan26B.
J.Biol.Chem., 294, 2019
|
|
3ICI
 
 | |
1VHN
 
 | |
