5T77
 
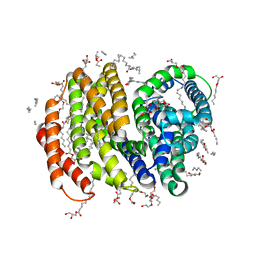 | | Crystal structure of the MOP flippase MurJ | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, ... | | Authors: | Kuk, A.C.Y, Lee, S.-Y. | | Deposit date: | 2016-09-02 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the MOP flippase MurJ in an inward-facing conformation.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6OO7
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in nanodiscs | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
6OB6
 
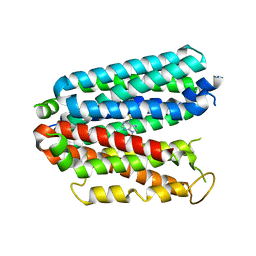 | |
6OB7
 
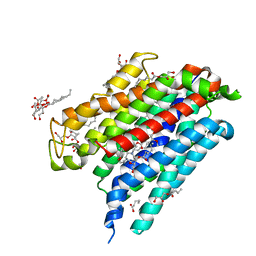 | | Human equilibrative nucleoside transporter-1, dilazep bound | | Descriptor: | (1,4-diazepane-1,4-diyl)di(propane-3,1-diyl) bis(3,4,5-trimethoxybenzoate), (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Equilibrative nucleoside transporter 1 | | Authors: | Wright, N.J, Lee, S.-Y. | | Deposit date: | 2019-03-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of human ENT1 in complex with adenosine reuptake inhibitors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6OO3
 
 | | Cryo-EM structure of the C4-symmetric TRPV2/RTx complex in amphipol resolved to 2.9 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
7RQZ
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 48 degrees Celsius, in an open state, class alpha | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQY
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 25 degrees Celsius, in an open state, class B | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
6OO4
 
 | | Cryo-EM structure of the C2-symmetric TRPV2/RTx complex in amphipol resolved to 3.3 A | | Descriptor: | TRPV2, resiniferatoxin | | Authors: | Zubcevic, L, Hsu, A.L, Borgnia, M.J, Lee, S.-Y. | | Deposit date: | 2019-04-22 | | Release date: | 2019-05-29 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Symmetry transitions during gating of the TRPV2 ion channel in lipid membranes.
Elife, 8, 2019
|
|
7RQX
 
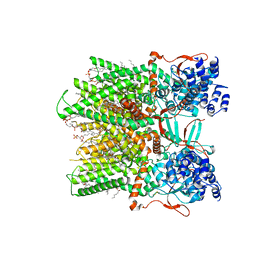 | | Cryo-EM structure of the full-length TRPV1 with RTx at 25 degrees Celsius, in an intermediate-open state, class A | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQU
 
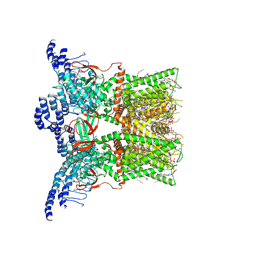 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in a closed state, class I | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, SODIUM ION, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQW
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in an open state, class III | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQV
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in an intermediate-closed state, class II | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
6PKW
 
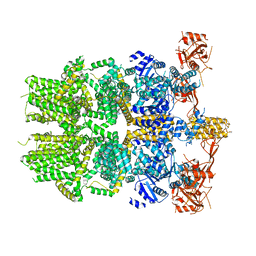 | | Cryo-EM structure of the zebrafish TRPM2 channel in the apo conformation, processed with C2 symmetry (pseudo C4 symmetry) | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
6PKV
 
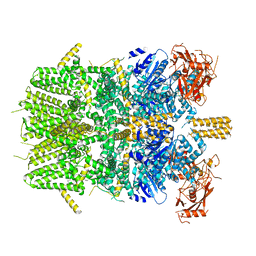 | | Cryo-EM structure of the zebrafish TRPM2 channel in the apo conformation, processed with C4 symmetry | | Descriptor: | Transient receptor potential cation channel subfamily M member 2 | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
6PQP
 
 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist BITC | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-benzylthioformamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
6PKX
 
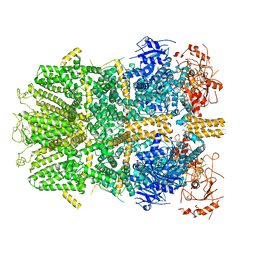 | | Cryo-EM structure of the zebrafish TRPM2 channel in the presence of ADPR and Ca2+ | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily M member 2, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Yin, Y, Wu, M, Hsu, A.L, Borschel, W.F, Borgnia, M.J, Lander, G.C, Lee, S.-Y. | | Deposit date: | 2019-06-30 | | Release date: | 2019-08-28 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Visualizing structural transitions of ligand-dependent gating of the TRPM2 channel.
Nat Commun, 10, 2019
|
|
6PQQ
 
 | | Cryo-EM structure of human TRPA1 C621S mutant in the apo state | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily A member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
6PQO
 
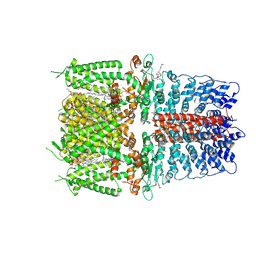 | | Cryo-EM structure of the human TRPA1 ion channel in complex with the covalent agonist JT010 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-N-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-N-(3-methoxypropyl)acetamide, ... | | Authors: | Suo, Y, Wang, Z, Zubcevic, L, Hsu, A.L, He, Q, Borgnia, M.J, Ji, R.-R, Lee, S.-Y. | | Deposit date: | 2019-07-09 | | Release date: | 2020-01-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel.
Neuron, 105, 2020
|
|
8TZL
 
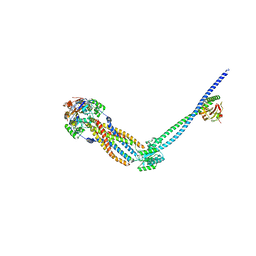 | | Cryo-EM structure of Vibrio cholerae FtsE/FtsX/EnvC complex, full-length | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural insights into the FtsEX-EnvC complex regulation on septal peptidoglycan hydrolysis in Vibrio cholerae.
Structure, 32, 2024
|
|
8TZK
 
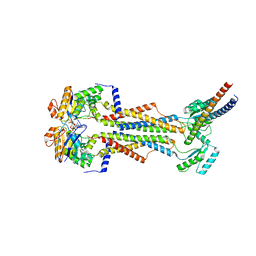 | | Cryo-EM structure of Vibrio cholerae FtsE/FtsX/EnvC complex, shortened | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural insights into the FtsEX-EnvC complex regulation on septal peptidoglycan hydrolysis in Vibrio cholerae.
Structure, 32, 2024
|
|
8TZJ
 
 | | Cryo-EM structure of Vibrio cholerae FtsE/FtsX complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE, Cell division protein FtsX, ... | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2023-08-27 | | Release date: | 2023-12-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural insights into the FtsEX-EnvC complex regulation on septal peptidoglycan hydrolysis in Vibrio cholerae.
Structure, 32, 2024
|
|
8TZ6
 
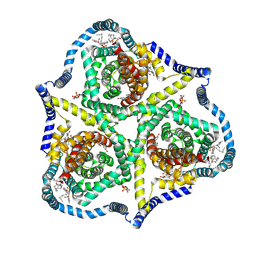 | |
8TZ8
 
 | |
8TZ9
 
 | |
8TZ1
 
 | |
