8H00
 
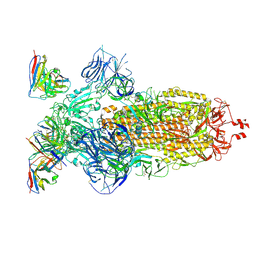 | | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8HEC
 
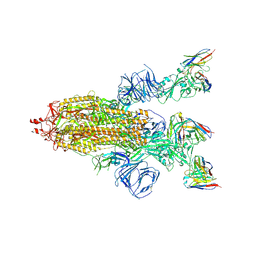 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
7LIO
 
 | |
7LIQ
 
 | |
8GZZ
 
 | | Local refinement of SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab | | Descriptor: | Spike protein S1, rabbit monoclonal antibody 1H1 Fab heavy chain, rabbit monoclonal antibody 1H1 Fab light chain | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-09-27 | | Release date: | 2023-04-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants.
Commun Biol, 6, 2023
|
|
8HED
 
 | | Local refinement of the SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.59 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
7LIP
 
 | | X-ray structure of SPOP MATH domain (D140G) | | Descriptor: | SULFATE ION, Speckle-type POZ protein | | Authors: | Botuyan, M.V, Cui, G, Mer, G. | | Deposit date: | 2021-01-27 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | ATM-phosphorylated SPOP contributes to 53BP1 exclusion from chromatin during DNA replication.
Sci Adv, 7, 2021
|
|
7LIN
 
 | |
8HEB
 
 | | SARS-CoV-2 Spike trimer in complex with RmAb 9H1 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-11-08 | | Release date: | 2023-04-26 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Mechanism of an RBM-targeted rabbit monoclonal antibody 9H1 neutralizing SARS-CoV-2.
Biochem.Biophys.Res.Commun., 660, 2023
|
|
5J39
 
 | | Crystal Structure of the extended TUDOR domain from TDRD2 | | Descriptor: | CACODYLATE ION, Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | Authors: | Zhang, H, Tempel, W, Dong, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-03-30 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7UZN
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX WITH BMT-206059 AKA 2-{(3M)-3-(1,4-DIMETHYL-1H-1,2,3-TRIAZOL-5-YL)-8-FLUORO-5-[(S)-(OXAN-4-YL)(PHENYL)METHYL]-5H-PYRIDO[3,2-b]INDOL-7-YL}PROPAN-2-OL, TRIPLY DEUTERATED ON THE 4-METHYL GROUP | | Descriptor: | 1,2-ETHANEDIOL, 2-{(3M)-3-(1,4-dimethyl-1H-1,2,3-triazol-5-yl)-8-fluoro-5-[(S)-(oxan-4-yl)(phenyl)methyl]-5H-pyrido[3,2-b]indol-7-yl}propan-2-ol, Bromodomain-containing protein 4, ... | | Authors: | Sheriff, S. | | Deposit date: | 2022-05-09 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.685 Å) | | Cite: | Development of BET Inhibitors as Potential Treatments for Cancer: Optimization of Pharmacokinetic Properties.
Acs Med.Chem.Lett., 13, 2022
|
|
6C4Y
 
 | | Cross-alpha Amyloid-like Structure alphaAmG | | Descriptor: | Cross-alpha Amyloid-like Structure alphaAmG | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
5HJB
 
 | | AF9 YEATS in complex with histone H3 Crotonylation at K9 | | Descriptor: | Protein AF-9, peptide of Histone H3.1 | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-12 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
5HJD
 
 | | AF9 YEATS in complex with histone H3 Crotonylation at K18 | | Descriptor: | COPPER (II) ION, Protein AF-9, SULFATE ION, ... | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain.
Mol.Cell, 62, 2016
|
|
6DIG
 
 | | Crystal structure of DQA1*01:02/DQB1*06:02 in complex with a hypocretin peptide | | Descriptor: | 13-mer peptide: ALA-GLY-ASN-HIS-ALA-ALA-GLY-ILE-LEU-THR-LEU-GLY-LYS, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Birtley, J.R, Stern, L.J, Mellins, E.D, Jiang, W. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vivo clonal expansion and phenotypes of hypocretin-specific CD4+T cells in narcolepsy patients and controls.
Nat Commun, 10, 2019
|
|
5HJC
 
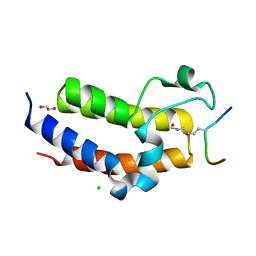 | | BRD3 second bromodomain in complex with histone H3 acetylation at K18 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 3, CHLORIDE ION, ... | | Authors: | Li, Y.Y, Zhao, D, Guan, H.P, Li, H.T. | | Deposit date: | 2016-01-13 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain
Mol.Cell, 62, 2016
|
|
6C4X
 
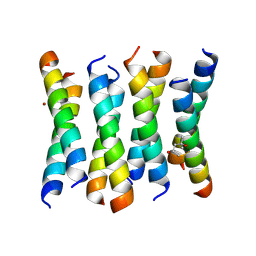 | | Cross-alpha Amyloid-like Structure alphaAmmem | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ZINC ION, cross-alpha amyloid-like membrane peptide alpha-AmMEM | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C50
 
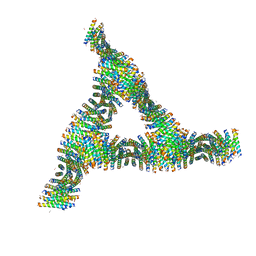 | | Cross-alpha Amyloid-like Structure alphaAmS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmS, FORMIC ACID | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C4Z
 
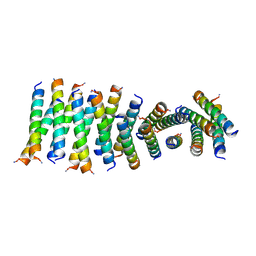 | |
2CMH
 
 | |
2CMG
 
 | |
6C52
 
 | | Cross-alpha Amyloid-like Structure alphaTet | | Descriptor: | Cross-alpha Amyloid-like Structure alphaTet, GLYCEROL | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6D02
 
 | | Cross-alpha Amyloid-like Structure alphaAmL, 2nd form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Zhang, S.-Q, Liu, L, Degrado, W.F. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
8FIU
 
 | | Potent long-acting inhibitors targeting HIV-1 capsid based on a versatile quinazolin-4-one scaffold | | Descriptor: | 1,2-ETHANEDIOL, HIV-1 capsid, N-[(1S)-1-{(3P)-3-{4-chloro-3-[(methanesulfonyl)amino]-1-methyl-1H-indazol-7-yl}-7-[(2R,6S)-2,6-dimethylmorpholin-4-yl]-4-oxo-3,4-dihydroquinazolin-2-yl}-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-3-(difluoromethyl)-5,5-difluoro-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Nolte, R.T. | | Deposit date: | 2022-12-16 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Potent Long-Acting Inhibitors Targeting the HIV-1 Capsid Based on a Versatile Quinazolin-4-one Scaffold.
J.Med.Chem., 66, 2023
|
|
6C51
 
 | | Cross-alpha Amyloid-like Structure alphaAmL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmL, PHOSPHATE ION | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
