3GGP
 
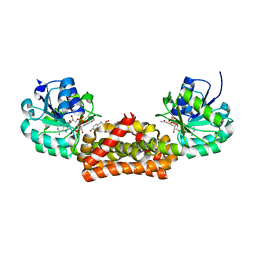 | | Crystal structure of prephenate dehydrogenase from A. aeolicus in complex with hydroxyphenyl propionate and NAD+ | | Descriptor: | CHLORIDE ION, HYDROXYPHENYL PROPIONIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Sun, W, Shahinas, D, Kimber, M.S, Christendat, D. | | Deposit date: | 2009-03-01 | | Release date: | 2009-03-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Crystal Structure of Aquifex aeolicus Prephenate Dehydrogenase Reveals the Mode of Tyrosine Inhibition.
J.Biol.Chem., 284, 2009
|
|
8PU7
 
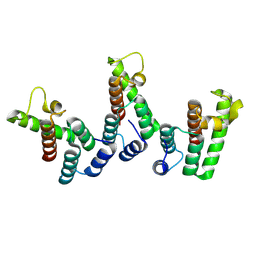 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -20 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PUA
 
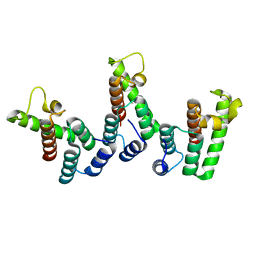 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -05 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PUB
 
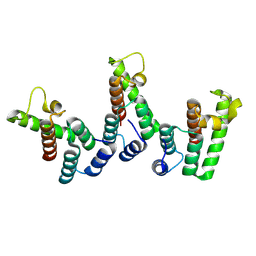 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 0 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PUD
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 10 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PUH
 
 | | Structure of the immature HTLV-1 CA lattice from full-length Gag VLPs: CA-CTD refinement | | Descriptor: | Gag polyprotein | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PU8
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle -15 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8PUE
 
 | | Structure of immature HTLV-1 CA-NTD from in vitro assembled MA126-CANC tubes: axis angle 15 degrees | | Descriptor: | Gag protein (Fragment) | | Authors: | Obr, M, Percipalle, M, Chernikova, D, Yang, H, Thader, A, Pinke, G, Porley, D, Mansky, L.M, Dick, R.A, Schur, F.K.M. | | Deposit date: | 2023-07-17 | | Release date: | 2023-08-23 | | Last modified: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Distinct stabilization of the human T cell leukemia virus type 1 immature Gag lattice.
Nat.Struct.Mol.Biol., 32, 2025
|
|
3G9E
 
 | | Aleglitaar. a new. potent, and balanced dual ppara/g agonist for the treatment of type II diabetes | | Descriptor: | (2S)-2-methoxy-3-{4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Ruf, A, Benz, J, Bernardeau, A, Binggeli, A, Blum, D, Boehringer, M, Grether, U, Hilpert, H, Kuhn, B, Maerki, H.P, Meyer, M, Puenterner, K, Raab, S, Schlatter, D, Gsell, B, Stihle, M, Mohr, P. | | Deposit date: | 2009-02-13 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Aleglitazar, a new, potent, and balanced dual PPARalpha/gamma agonist for the treatment of type II diabetes.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
4AFQ
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHYMASE, CITRATE ANION, ... | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
3GGG
 
 | |
1WRT
 
 | | NMR STUDY OF APO TRP REPRESSOR | | Descriptor: | APO TRP REPRESSOR | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
1CLE
 
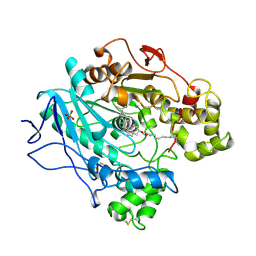 | | STRUCTURE OF UNCOMPLEXED AND LINOLEATE-BOUND CANDIDA CYLINDRACEA CHOLESTEROL ESTERASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL ESTERASE, ... | | Authors: | Ghosh, D. | | Deposit date: | 1995-02-08 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of uncomplexed and linoleate-bound Candida cylindracea cholesterol esterase.
Structure, 3, 1995
|
|
1WRS
 
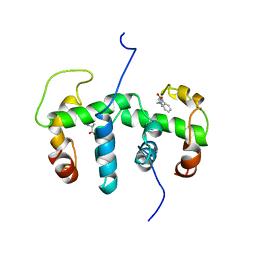 | | NMR STUDY OF HOLO TRP REPRESSOR | | Descriptor: | HOLO TRP REPRESSOR, TRYPTOPHAN | | Authors: | Zhao, D, Zheng, Z. | | Deposit date: | 1995-05-12 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structures of the Escherichia coli trp holo- and aporepressor.
J.Mol.Biol., 229, 1993
|
|
2VX0
 
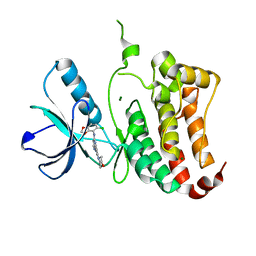 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N'-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-N-(3-MORPHOLIN-4-YLPHENYL)PYRIMIDINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-30 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4A0Y
 
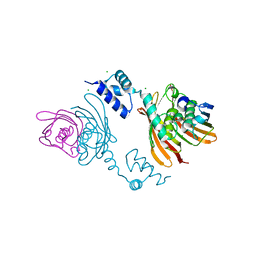 | | Structure of the global transcription regulator FapR from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, TRANSCRIPTION FACTOR FAPR | | Authors: | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | Deposit date: | 2011-09-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
4AP5
 
 | | Crystal structure of human POFUT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GDP-FUCOSE PROTEIN O-FUCOSYLTRANSFERASE 2, ... | | Authors: | Chen, C, Keusch, J.J, Klein, D, Hess, D, Hofsteenge, J, Gut, H. | | Deposit date: | 2012-03-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structure of Human Pofut2: Insights Into Thrombospondin Type 1 Repeat Fold and O-Fucosylation.
Embo J., 31, 2012
|
|
6GCH
 
 | | STRUCTURE OF CHYMOTRYPSIN-*TRIFLUOROMETHYL KETONE INHIBITOR COMPLEXES. COMPARISON OF SLOWLY AND RAPIDLY EQUILIBRATING INHIBITORS | | Descriptor: | 1,1,1-TRIFLUORO-3-ACETAMIDO-4-PHENYL BUTAN-2-ONE(N-ACETYL-L-PHENYLALANYL TRIFLUOROMETHYL KETONE), GAMMA-CHYMOTRYPSIN A | | Authors: | Brady, K, Wei, A, Ringe, D, Abeles, R.H. | | Deposit date: | 1990-04-06 | | Release date: | 1990-10-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of chymotrypsin-trifluoromethyl ketone inhibitor complexes: comparison of slowly and rapidly equilibrating inhibitors.
Biochemistry, 29, 1990
|
|
6SNW
 
 | | Structure of Coxsackievirus A10 complexed with its receptor KREMEN1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP3, ... | | Authors: | Zhao, Y, Zhou, D, Ni, T, Karia, D, Kotecha, A, Wang, X, Rao, Z, Jones, E.Y, Fry, E.E, Ren, J, Stuart, D.I. | | Deposit date: | 2019-08-27 | | Release date: | 2020-01-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10.
Nat Commun, 11, 2020
|
|
2VRN
 
 | | The structure of the stress response protein DR1199 from Deinococcus radiodurans: a member of the DJ-1 superfamily | | Descriptor: | MAGNESIUM ION, PROTEASE I | | Authors: | Fioravanti, E, Dura, M.A, Lascoux, D, Micossi, E, McSweeney, S. | | Deposit date: | 2008-04-09 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the stress response protein DR1199 from Deinococcus radiodurans: a member of the DJ-1 superfamily.
Biochemistry, 47, 2008
|
|
4AQY
 
 | | Structure of ribosome-apramycin complexes | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Matt, T, Ng, C.L, Lang, K, Sha, S.H, Akbergenov, R, Shcherbakov, D, Meyer, M, Duscha, S, Xie, J, Dubbaka, S.R, Perez-Fernandez, D, Vasella, A, Ramakrishnan, V, Schacht, J, Bottger, E.C. | | Deposit date: | 2012-04-20 | | Release date: | 2012-07-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Dissociation of Antibacterial Activity and Aminoglycoside Ototoxicity in the 4-Monosubstituted 2-Deoxystreptamine Apramycin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1D0E
 
 | | CRYSTAL STRUCTURES OF THE N-TERMINAL FRAGMENT FROM MOLONEY MURINE LEUKEMIA VIRUS REVERSE TRANSCRIPTASE COMPLEXED WITH NUCLEIC ACID: FUNCTIONAL IMPLICATIONS FOR TEMPLATE-PRIMER BINDING TO THE FINGERS DOMAIN | | Descriptor: | 5'-D(*TP*TP*TP*CP*AP*TP*GP*CP*AP*TP*G)-3', REVERSE TRANSCRIPTASE | | Authors: | Najmudin, S, Cote, M.L, Sun, D, Yohannan, S, Montano, S.P, Gu, J, Georgiadis, M.M. | | Deposit date: | 1999-09-09 | | Release date: | 2000-04-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template-primer binding to the fingers domain
J.Mol.Biol., 296, 2000
|
|
1ZK3
 
 | | Triclinic crystal structure of the apo-form of R-specific alcohol dehydrogenase (mutant G37D) from Lactobacillus brevis | | Descriptor: | MAGNESIUM ION, R-specific alcohol dehydrogenase | | Authors: | Schlieben, N.H, Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2005-05-02 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic Resolution Structures of R-specific Alcohol Dehydrogenase from Lactobacillus brevis Provide the Structural Bases of its Substrate and Cosubstrate Specificity
J.Mol.Biol., 349, 2005
|
|
6O4J
 
 | | Amyloid Beta KLVFFAENVGS 16-26 D23N Iowa mutation | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Griner, S.L, Sawaya, M.R, Rodriguez, J.A, Cascio, D, Gonen, T. | | Deposit date: | 2019-02-28 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.402 Å) | | Cite: | Structure based inhibitors of Amyloid Beta core suggest a common interface with Tau.
Elife, 8, 2019
|
|
8QV0
 
 | | Structure of the native microtubule lattice nucleated from the yeast spindle pole body | | Descriptor: | Tubulin alpha-1 chain, Tubulin beta chain | | Authors: | Dendooven, T, Yatskevich, S, Burt, A, Bellini, D, Kilmartin, J, Barford, D. | | Deposit date: | 2023-10-17 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structure of the native gamma-tubulin ring complex capping spindle microtubules.
Nat.Struct.Mol.Biol., 31, 2024
|
|
