7AT6
 
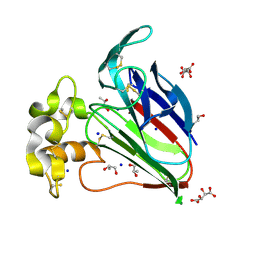 | | Structure of thaumatin collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | L(+)-TARTARIC ACID, R-1,2-PROPANEDIOL, SODIUM ION, ... | | Authors: | Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Wang, M, Pedrini, B. | | Deposit date: | 2020-10-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
5D5A
 
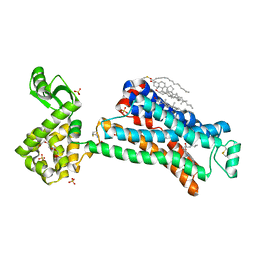 | | In meso in situ serial X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Liu, X, Kobilka, B, Kay Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4826 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3ZOL
 
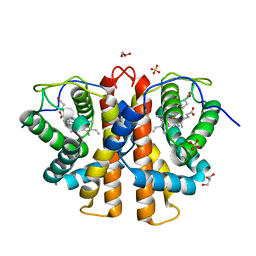 | | M.acetivorans protoglobin F93Y mutant in complex with cyanide | | Descriptor: | CYANIDE ION, GLYCEROL, PROTOGLOBIN, ... | | Authors: | Tilleman, L, Abbruzzetti, S, Ciaccio, C, De Sanctis, G, Nardini, M, Pesce, A, Desmet, F, Moens, L, Van Doorslaer, S, Bruno, S, Bolognesi, M, Ascenzi, P, Coletta, M, Viappiani, C, Dewilde, S. | | Deposit date: | 2013-02-22 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Bases for the Regulation of Co Binding in the Archaeal Protoglobin from Methanosarcina Acetivorans.
Plos One, 10, 2015
|
|
3EBL
 
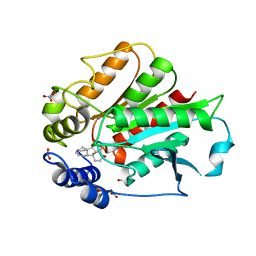 | | Crystal Structure of Rice GID1 complexed with GA4 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GIBBERELLIN A4, Gibberellin receptor GID1, ... | | Authors: | Shimada, A, Nakatsu, T, Ueguchi-Tanaka, M, Kato, H, Matsuoka, M. | | Deposit date: | 2008-08-28 | | Release date: | 2008-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for gibberellin recognition by its receptor GID1.
Nature, 456, 2008
|
|
7B1T
 
 | | Crystal structure of BRD4(1) in complex with the inhibitor MPM6 | | Descriptor: | 3-(5-azanyl-2-chloranyl-phenyl)-1-methyl-4,7-dihydro-2~{H}-cyclohepta[c]pyrrol-8-one, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE | | Authors: | Huegle, M. | | Deposit date: | 2020-11-25 | | Release date: | 2022-06-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A novel pan-selective bromodomain inhibitor for epigenetic drug design.
Eur.J.Med.Chem., 249, 2023
|
|
3ZJH
 
 | | Trp(60)B9Ala mutation of M.acetivorans protoglobin in complex with cyanide | | Descriptor: | CYANIDE ION, PROTOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
5D53
 
 | | In meso in situ serial X-ray crystallography structure of insulin at 100 K | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, Insulin A chain, Insulin B chain, ... | | Authors: | Huang, C.-Y, Olieric, V, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D56
 
 | | In meso in situ serial X-ray crystallography structure of diacylglycerol kinase, DgkA, at 100 K | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, ACETATE ION, CITRATE ANION, ... | | Authors: | Huang, C.-Y, Howe, N, Olieric, V, Warshamanage, R, Diederichs, K, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5D5B
 
 | | In meso X-ray crystallography structure of the Beta2-adrenergic receptor at 100 K | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, 1,4-BUTANEDIOL, ACETAMIDE, ... | | Authors: | Huang, C.-Y, Olieric, V, Liu, X, Kobilka, B, Wang, M, Caffrey, M. | | Deposit date: | 2015-08-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
3ESX
 
 | | E16KE61KD126KD150K Flavodoxin from Anabaena | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Herguedas, B, Hermoso, J.A, Martinez-Julvez, M, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
3ZNR
 
 | | HDAC7 bound with inhibitor TMP269 | | Descriptor: | HISTONE DEACETYLASE 7, N-{[4-(4-phenyl-1,3-thiazol-2-yl)tetrahydro-2H-pyran-4-yl]methyl}-3-[5-(trifluoromethyl)-1,2,4-oxadiazol-3-yl]benzamide, POTASSIUM ION, ... | | Authors: | Lobera, m, madauss, k, pohlhaus, d, trump, r, nolan, m. | | Deposit date: | 2013-02-15 | | Release date: | 2013-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective Class Iia Histone Deacetylase Inhibition Via a Non-Chelating Zinc Binding Group
Nat.Chem.Biol., 9, 2013
|
|
1TJB
 
 | | Crystal Structure of a High Affinity Lanthanide-Binding Peptide (LBT) | | Descriptor: | CHLORIDE ION, Lanthanide-Binding Peptide, TERBIUM(III) ION | | Authors: | Nitz, M, Sherawat, M, Franz, K.J, Peisach, E, Allen, K.N, Imperiali, B. | | Deposit date: | 2004-06-03 | | Release date: | 2004-08-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Origin of the High Affinity of a Chemically Evolved Lanthanide-Binding Peptide
Angew.Chem.Int.Ed.Engl., 43, 2004
|
|
3ESY
 
 | | E16KE61K Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
3WNN
 
 | | D308A mutant of Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase complexed with isomaltooctaose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Cycloisomaltooligosaccharide glucanotransferase, ... | | Authors: | Suzuki, N, Fujimoto, Z, Kim, Y.M, Momma, M, Kishine, N, Suzuki, R, Suzuki, S, Kitamura, S, Kobayashi, M, Kimura, A, Funane, K. | | Deposit date: | 2013-12-10 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural elucidation of the cyclization mechanism of alpha-1,6-glucan by Bacillus circulans T-3040 cycloisomaltooligosaccharide glucanotransferase.
J.Biol.Chem., 289, 2014
|
|
3EOC
 
 | | Cdk2/CyclinA complexed with a imidazo triazin-2-amine | | Descriptor: | 5-methyl-7-phenyl-N-(3,4,5-trimethoxyphenyl)imidazo[5,1-f][1,2,4]triazin-2-amine, Cell division protein kinase 2, Cyclin-A2 | | Authors: | Cheung, M, Kuntz, K, Pobanz, M, Salovich, J, Wilson, B, Andrews, W, Shewchuk, L, Epperly, A, Hassler, D, Leesnitzer, M, Smith, J, Smith, G, Lansing, T, Mook, R. | | Deposit date: | 2008-09-26 | | Release date: | 2008-11-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Imidazo[5,1-f][1,2,4]triazin-2-amines as novel inhibitors of polo-like kinase 1.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3WU4
 
 | | Oxidized-form structure of E.coli Lon Proteolytic domain | | Descriptor: | Lon protease, SULFATE ION | | Authors: | Nishii, W, Kukimoto-Niino, M, Terada, T, Shirouzu, M, Muramatsu, T, Yokoyama, S. | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A redox switch shapes the Lon protease exit pore to facultatively regulate proteolysis.
Nat. Chem. Biol., 11, 2015
|
|
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K54
 
 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in active R-state | | Descriptor: | Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
4ILC
 
 | | The GLIC pentameric ligand-gated ion channel in complex with sulfates | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Malherbe, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
8IEG
 
 | | Bre1(mRBD-RING)/Rad6-Ub/nucleosome complex | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase BRE1, Histone H2A type 1-B/E, ... | | Authors: | Ai, H, Deng, Z, Pan, M, Liu, L. | | Deposit date: | 2023-02-15 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanistic insights into nucleosomal H2B monoubiquitylation mediated by yeast Bre1-Rad6 and its human homolog RNF20/RNF40-hRAD6A.
Mol.Cell, 83, 2023
|
|
4II4
 
 | |
6IP8
 
 | | Cryo-EM structure of the HCV IRES dependently initiated CMV-stalled 80S ribosome (Structure iv) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
8I0E
 
 | | Sb3GT1 complex with UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Wang, H.T, Wang, Z.L, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
4IIT
 
 | |
8HZZ
 
 | | GuApiGT (UGT79B74) | | Descriptor: | SULFATE ION, apiosyltransferase | | Authors: | Wang, H.T, Wang, Z.L, Li, F.D, Ye, M. | | Deposit date: | 2023-01-10 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the missing apiosylation step in flavonoid apiosides biosynthesis of Leguminosae plants.
Nat Commun, 14, 2023
|
|
