1MAF
 
 | |
1MAE
 
 | |
1PVU
 
 | |
1PKP
 
 | |
1QGD
 
 | |
1QAW
 
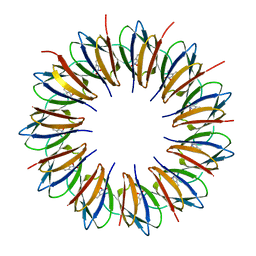 | | Regulatory Features of the TRP Operon and the Crystal Structure of the TRP RNA-Binding Attenuation Protein from Bacillus Stearothermophilus. | | Descriptor: | TRP RNA-BINDING ATTENUATION PROTEIN, TRYPTOPHAN | | Authors: | Chen, X.-P, Antson, A.A, Yang, M, Baumann, C, Dodson, E.J, Dodson, G.G, Gollnick, P. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus.
J.Mol.Biol., 289, 1999
|
|
1XYZ
 
 | |
1XKJ
 
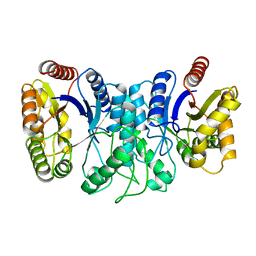 | | BACTERIAL LUCIFERASE BETA2 HOMODIMER | | Descriptor: | BETA2 LUCIFERASE | | Authors: | Tanner, J.J, Krause, K.L. | | Deposit date: | 1996-10-08 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of bacterial luciferase beta 2 homodimer: implications for flavin binding.
Biochemistry, 36, 1997
|
|
1GMQ
 
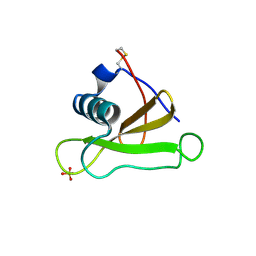 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1GMP
 
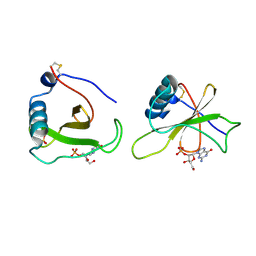 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1GMR
 
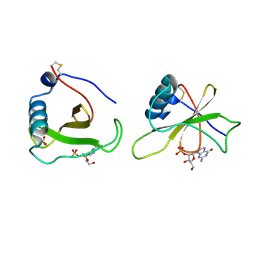 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
3TUF
 
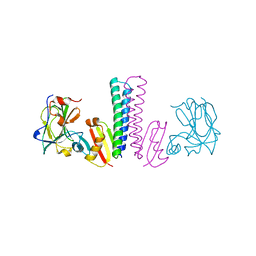 | |
2MAD
 
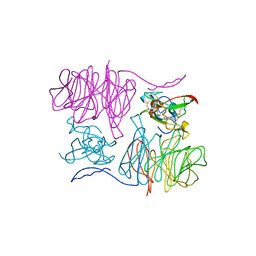 | |
2NLR
 
 | |
7QI3
 
 | | Structure of Fusarium verticillioides NAT1 (FDB2) N-malonyltransferase | | Descriptor: | 1,2-ETHANEDIOL, Arylamine N-acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lowe, E.D, Kotomina, E, Karagianni, E, Boukouvala, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fusarium verticillioides NAT1 (FDB2) N-malonyltransferase is structurally, functionally and phylogenetically distinct from its N-acetyltransferase (NAT) homologues.
Febs J., 290, 2023
|
|
2PRD
 
 | |
2OXI
 
 | |
6Q7J
 
 | | GH3 exo-beta-xylosidase (XlnD) in complex with xylobiose aziridine activity based probe | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6QE8
 
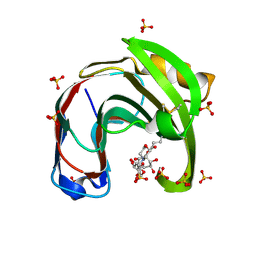 | | Crystal structure of Aspergillus niger GH11 endoxylanase XynA in complex with xylobiose epoxide activity based probe | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-5-[(2~{S},3~{R},4~{S},5~{R})-3,4,5-tris(oxidanyl)oxan-2-yl]oxycyclohexane-1,2,3,4-tetrol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase A, ... | | Authors: | Wu, L, Rowland, R.J, Davies, G.J. | | Deposit date: | 2019-01-07 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q7I
 
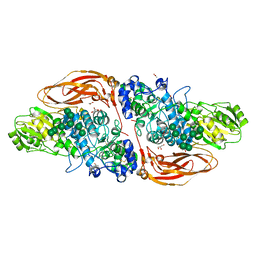 | | GH3 exo-beta-xylosidase (XlnD) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q8N
 
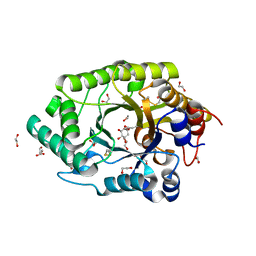 | | GH10 endo-xylanase in complex with xylobiose epoxide inhibitor | | Descriptor: | (1~{R},2~{S},4~{S},5~{R})-cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
6Q8M
 
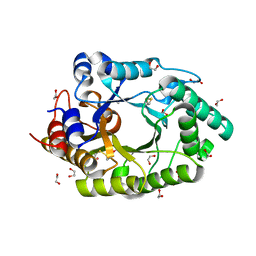 | | GH10 endo-xylanase | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-xylanase, ... | | Authors: | Davies, G.J, Rowland, R.J, Wu, L, Moroz, O, Blagova, E. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Dynamic and Functional Profiling of Xylan-Degrading Enzymes inAspergillusSecretomes Using Activity-Based Probes.
Acs Cent.Sci., 5, 2019
|
|
2J63
 
 | | Crystal structure of AP endonuclease LMAP from Leishmania major | | Descriptor: | AP-ENDONUCLEASE | | Authors: | Vidal, A.E, Harkiolaki, M, Gallego, C, Castillo-Acosta, V.M, Ruiz-Perez, L.M, Wilson, K.S, Gonzalez-Pacanowska, D. | | Deposit date: | 2006-09-25 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure and DNA Repair Activities of the Ap Endonuclease from Leishmania Major.
J.Mol.Biol., 373, 2007
|
|
1OBG
 
 | | SAICAR-synthase complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, PHOSPHORIBOSYLAMIDOIMIDAZOLE- SUCCINOCARBOXAMIDE SYNTHASE, ... | | Authors: | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
1OBD
 
 | | SAICAR-synthase complexed with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Antonyuk, S.V, Grebenko, A.I, Levdikov, V.M, Urusova, D.V, Melik-Adamyan, W.R, Lamzin, V.S, Wilson, K. | | Deposit date: | 2003-01-30 | | Release date: | 2003-03-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-Ray Structure of Saicar-Synthase Complexed with ATP
Kristallografiya, 46, 2001
|
|
