1IS8
 
 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex plus Zn | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1VTG
 
 | | THE MOLECULAR STRUCTURE OF A DNA-TRIOSTIN A COMPLEX | | Descriptor: | 2-CARBOXYQUINOXALINE, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3'), triostin A | | Authors: | Wang, A.H.-J, Ughetto, G, Quigley, G.J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The molecular structure of a DNA-triostin A complex.
Science, 225, 1984
|
|
1VTW
 
 | | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of D(M(5)CGTAM(5)CG) | | Descriptor: | DNA (5'-D(*(CH3)CP*GP*TP*AP*(CH3)CP*G)-3') | | Authors: | Wang, A.H.-J, Hakoshima, T, Van Der Marel, G.A, Van Boom, J.H, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | AT Base Pairs Are Less Stable than GC Base Pairs in Z-DNA: The Crystal Structure of d(m(5)CGTAm(5)CG)
Cell(Cambridge,Mass.), 37, 1984
|
|
3A9W
 
 | | Crystal structure of L-Threonine bound L-Threonine dehydrogenase (Y137F) from Hyperthermophilic Archaeon Thermoplasma volcanium | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, NDP-sugar epimerase, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2009-11-07 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Binary and Ternary Complexes of Archaeal UDP-galactose 4-Epimerase-like L-Threonine Dehydrogenase from Thermoplasma volcanium.
J.Biol.Chem., 287, 2012
|
|
3A4V
 
 | | Crystal structure of pyruvate bound L-Threonine dehydrogenase from Hyperthermophilic Archaeon Thermoplasma volcanium | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, NDP-sugar epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2009-07-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Binary and Ternary Complexes of Archaeal UDP-galactose 4-Epimerase-like L-Threonine Dehydrogenase from Thermoplasma volcanium.
J.Biol.Chem., 287, 2012
|
|
3AIB
 
 | | Crystal Structure of Glucansucrase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3WZ0
 
 | | On archaeal homologs of the human RNase P proteins Pop5 and Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis | | Descriptor: | Ribonuclease P protein component 2, Ribonuclease P protein component 3 | | Authors: | Suematsu, K, Ueda, T, Nakashima, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | On archaeal homologs of the human RNase P proteins Pop5 and Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
3AIE
 
 | | Crystal Structure of glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Glucosyltransferase-SI | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
3WYZ
 
 | | On archaeal homologs of the human RNase P protein Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis | | Descriptor: | GLYCEROL, Ribonuclease P protein component 3 | | Authors: | Suematsu, K, Ueda, T, Nakashima, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2014-09-11 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | On archaeal homologs of the human RNase P proteins Pop5 and Rpp30 in the hyperthermophilic archaeon Thermococcus kodakarensis.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
3AIC
 
 | | Crystal Structure of Glucansucrase from Streptococcus mutans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ito, K, Ito, S, Shimamura, T, Iwata, S. | | Deposit date: | 2010-05-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of glucansucrase from the dental caries pathogen Streptococcus mutans.
J.Mol.Biol., 408, 2011
|
|
7YA8
 
 | | The crystal structure of IpaH2.5 LRR domain | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Hiragi, K, Nishide, A, Takagi, K, Iwai, K, Kim, M, Mizushima, T. | | Deposit date: | 2022-06-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
J.Biochem., 173, 2023
|
|
3AJR
 
 | | Crystal structure of L-3-Hydroxynorvaline bound L-Threonine dehydrogenase (Y137F) from Hyperthermophilic Archaeon Thermoplasma volcanium | | Descriptor: | (3R)-3-hydroxy-L-norvaline, (4S)-2-METHYL-2,4-PENTANEDIOL, NDP-sugar epimerase, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2010-06-14 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal Structure of Binary and Ternary Complexes of Archaeal UDP-galactose 4-Epimerase-like L-Threonine Dehydrogenase from Thermoplasma volcanium.
J.Biol.Chem., 287, 2012
|
|
7YA7
 
 | | The crystal structure of IpaH1.4 LRR domain | | Descriptor: | RING-type E3 ubiquitin transferase | | Authors: | Hiragi, K, Nishide, A, Takagi, K, Iwai, K, Kim, M, Mizushima, T. | | Deposit date: | 2022-06-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insight into the recognition of the linear ubiquitin assembly complex by Shigella E3 ligase IpaH1.4/2.5.
J.Biochem., 173, 2023
|
|
3A1N
 
 | |
3VPX
 
 | | Crystal structure of leucine dehydrogenase from a psychrophilic bacterium Sporosarcina psychrophila. | | Descriptor: | Leucine dehydrogenase | | Authors: | Zhao, Y, Wakamatsu, T, Doi, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2012-03-14 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A psychrophilic leucine dehydrogenase from Sporosarcina psychrophila: Purification, characterization, gene sequencing and crystal structure analysis
J.MOL.CATAL., B ENZYM., 83, 2012
|
|
2ZQD
 
 | | Ceftazidime acyl-intermediate structure of class a beta-lact Toho-1 E166A/R274N/R276N triple mutant | | Descriptor: | ACYLATED CEFTAZIDIME, Beta-lactamase Toho-1, SULFATE ION | | Authors: | Nitanai, Y, Shimamura, T, Uchiyama, T, Ishii, Y, Takehira, M, Yutani, K, Matsuzawa, H, Miyano, M. | | Deposit date: | 2008-08-07 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Structural Basis of Extend Spectrum Beta-Lactamase in Correlation of Enzymatic Kinetics and Thermal Stability of Acyl-Intermediates
To be Published
|
|
3VQR
 
 | | Structure of a dye-linked L-proline dehydrogenase mutant from the aerobic hyperthermophilic archaeon, Aeropyrum pernix | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sakuraba, H, Ohshima, T, Satomura, T, Yoneda, K. | | Deposit date: | 2012-03-29 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal Structure of Novel Dye-linked L-Proline Dehydrogenase from Hyperthermophilic Archaeon Aeropyrum pernix
J.Biol.Chem., 287, 2012
|
|
5XFL
 
 | |
3A7S
 
 | | Catalytic domain of UCH37 | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Nishio, K, Kim, S.W, Kawai, K, Mizushima, T, Yamane, T, Hamazaki, J, Murata, S, Tanaka, K. | | Deposit date: | 2009-10-04 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the de-ubiquitinating enzyme UCH37 (human UCH-L5) catalytic domain
Biochem.Biophys.Res.Commun., 2009
|
|
3W06
 
 | | Crystal structure of Arabidopsis thaliana DWARF14 Like (AtD14L) | | Descriptor: | 1,2-ETHANEDIOL, Hydrolase, alpha/beta fold family protein | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3W04
 
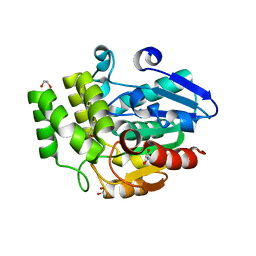 | | Crystal structure of Oryza sativa DWARF14 (D14) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Kagiyama, M, Hirano, Y, Mori, T, Kim, S.Y, Kyozuka, J, Seto, Y, Yamaguchi, S, Hakoshima, T. | | Deposit date: | 2012-10-19 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of D14 and D14L in the strigolactone and karrikin signaling pathways
Genes Cells, 18, 2013
|
|
3ACP
 
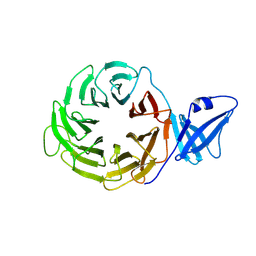 | | Crystal Structure of Yeast Rpn14, a Chaperone of the 19S Regulatory Particle of the Proteasome | | Descriptor: | WD repeat-containing protein YGL004C | | Authors: | Kim, S, Saeki, Y, Suzuki, A, Takagi, K, Fukunaga, K, Yamane, T, Kato, K, Tanaka, K, Mizushima, T. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of yeast Rpn14, a chaperone of the 19S regulatory particle of the proteasome
J.Biol.Chem., 285, 2010
|
|
3WRJ
 
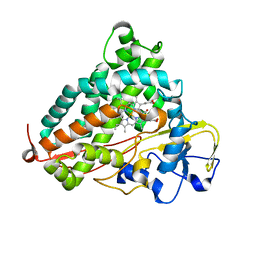 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of P450cam intermedite
to be published
|
|
3AU4
 
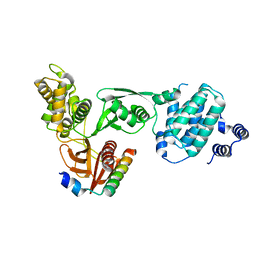 | |
3AW5
 
 | | Structure of a multicopper oxidase from the hyperthermophilic archaeon Pyrobaculum aerophilum | | Descriptor: | ACETATE ION, COPPER (II) ION, CU-O-CU LINKAGE, ... | | Authors: | Sakuraba, H, Ohshima, T, Yoneda, K. | | Deposit date: | 2011-03-10 | | Release date: | 2011-06-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a multicopper oxidase from the hyperthermophilic archaeon Pyrobaculum aerophilum
Acta Crystallogr.,Sect.F, 67, 2011
|
|
