3M9Y
 
 | | Crystal structure of Triosephosphate isomerase from methicillin resistant Staphylococcus aureus at 1.9 Angstrom resolution | | Descriptor: | CITRIC ACID, SODIUM ION, Triosephosphate isomerase | | Authors: | Mukherjee, S, Dutta, D, Saha, B, Das, A.K. | | Deposit date: | 2010-03-23 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of triosephosphate isomerase from methicillin resistant Staphylococcus aureus MRSA252 provide structural insights into novel modes of ligand binding and unique conformations of catalytic loop
Biochimie, 94, 2012
|
|
2QPQ
 
 | | Structure of Bug27 from Bordetella pertussis | | Descriptor: | CITRIC ACID, protein Bug27 | | Authors: | Herrou, J, Bompard, C. | | Deposit date: | 2007-07-25 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-based mechanism of ligand binding for periplasmic solute-binding protein of the Bug family.
J.Mol.Biol., 373, 2007
|
|
7PUF
 
 | | urate oxidase azaxanthine complex under 600 bar (60 MPa) of argon | | Descriptor: | 8-AZAXANTHINE, ARGON, SODIUM ION, ... | | Authors: | Prange, T, Colloc'h, N, Carpentier, P. | | Deposit date: | 2021-09-29 | | Release date: | 2022-02-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Comparative study of the effects of high hydrostatic pressure per se and high argon pressure on urate oxidase ligand stabilization
Acta Cryst. D, 78, 2022
|
|
6GC9
 
 | |
6GCA
 
 | |
6GCC
 
 | | Crystal structure of glutathione transferase Xi 3 mutant C56S from Trametes versicolor in complex with dextran-sulfate | | Descriptor: | 3,4-di-O-sulfo-alpha-D-glucopyranose-(1-6)-3,4-di-O-sulfo-alpha-D-altropyranose-(1-6)-1,2,3,4-tetra-O-sulfo-alpha-D-allopyranose, Glutathione transferase Xi 3 mutant C56S | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-04-17 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
FEBS Lett., 592, 2018
|
|
6HTA
 
 | | Crystal structure of glutathione transferase Xi 3 mutant C56S from Trametes versicolor | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, Glutathione transferase Xi 3 C56S | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Trametes versicolor glutathione transferase Xi 3, a dual Cys-GST with catalytic specificities of both Xi and Omega classes.
FEBS Lett., 592, 2018
|
|
4H35
 
 | |
3IMX
 
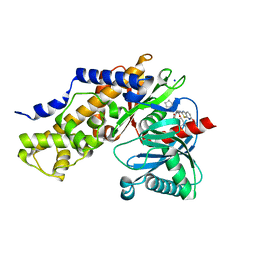 | | Crystal Structure of human glucokinase in complex with a synthetic activator | | Descriptor: | (2R)-3-cyclopentyl-N-(5-methoxy[1,3]thiazolo[5,4-b]pyridin-2-yl)-2-{4-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Stams, T, Vash, B. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of functionally liver selective glucokinase activators for the treatment of type 2 diabetes.
J.Med.Chem., 52, 2009
|
|
6GCB
 
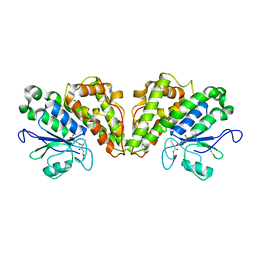 | |
4FW8
 
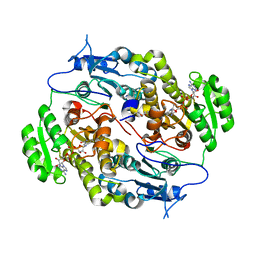 | | Crystal structure of FABG4 complexed with Coenzyme NADH | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-(Acyl-carrier-protein) reductase | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2012-06-30 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis
Biochem.J., 450, 2013
|
|
4N8D
 
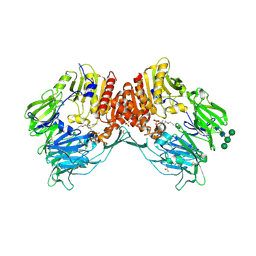 | | DPP4 complexed with syn-7aa | | Descriptor: | 1-(cis-1-phenyl-4-{[(2E)-3-phenylprop-2-en-1-yl]oxy}cyclohexyl)methanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of C-(1-aryl-cyclohexyl)-methylamines as selective, orally available inhibitors of dipeptidyl peptidase IV.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
4N8E
 
 | | DPP4 complexed with compound 12a | | Descriptor: | 1-[cis-4-(aminomethyl)-4-(3-chlorophenyl)cyclohexyl]piperidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, ... | | Authors: | Ostermann, N, Zink, F, Kroemer, M. | | Deposit date: | 2013-10-17 | | Release date: | 2014-02-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of C-(1-aryl-cyclohexyl)-methylamines as selective, orally available inhibitors of dipeptidyl peptidase IV.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3V1U
 
 | | Crystal structure of a beta-ketoacyl reductase FabG4 from Mycobacterium tuberculosis H37Rv complexed with NAD+ and Hexanoyl-CoA at 2.5 Angstrom resolution | | Descriptor: | (2S,5R,8R,11S,14S,17S,21R)-5,8,11,14,17-PENTAMETHYL-4,7,10,13,16,19-HEXAOXADOCOSANE-2,21-DIOL, 3-oxoacyl-(Acyl-carrier-protein) reductase, HEXANOYL-COENZYME A, ... | | Authors: | Dutta, D, Bhattacharyya, S, Das, A.K. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hexanoyl-CoA bound to beta-ketoacyl reductase FabG4 of Mycobacterium tuberculosis
Biochem.J., 450, 2013
|
|
4P78
 
 | | HicA3 and HicB3 toxin-antitoxin complex | | Descriptor: | GLYCEROL, HicA3 Toxin, HicB3 antitoxin | | Authors: | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | Deposit date: | 2014-03-26 | | Release date: | 2014-08-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
4P7D
 
 | | Antitoxin HicB3 crystal structure | | Descriptor: | Antitoxin HicB3, CHLORIDE ION | | Authors: | Li de la Sierra-Gallay, I, Bibi-Triki, S, van Tilbeurgh, H, Lazar, N, Pradel, E. | | Deposit date: | 2014-03-27 | | Release date: | 2014-08-27 | | Last modified: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Functional and Structural Analysis of HicA3-HicB3, a Novel Toxin-Antitoxin System of Yersinia pestis.
J.Bacteriol., 196, 2014
|
|
4ALT
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4BAG
 
 | |
4ALS
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALR
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALC
 
 | | X-Ray photoreduction of Polysaccharide monooxigenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALE
 
 | | Structure changes of Polysaccharide monooxygenase CBM33A from Enterococcus faecalis by X-ray induced photoreduction. | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-02 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4ALQ
 
 | | X-Ray photoreduction of Polysaccharide monooxygenase CBM33 | | Descriptor: | CHITIN BINDING PROTEIN, COPPER (II) ION, DI(HYDROXYETHYL)ETHER | | Authors: | Gudmundsson, M, Wu, M, Ishida, T, Momeni, M.H, Vaaje-Kolstad, G, Eijsink, V, Sandgren, M. | | Deposit date: | 2012-03-05 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and Electronic Snapshots During the Transition from a Cu(II) to Cu(I) Metal Center of a Lytic Polysaccharide Monooxygenase by X-Ray Photo-Reduction.
J.Biol.Chem., 289, 2014
|
|
4CW3
 
 | | Crystal structure of cofactor-free urate oxidase in complex with the 5-peroxo derivative of 9-metyl uric acid (X-ray dose, 665 kGy) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5S)-5-(dioxidanyl)-9-methyl-7H-purine-2,6,8-trione, 9-METHYL URIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-29 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Direct evidence for a peroxide intermediate and a reactive enzyme-substrate-dioxygen configuration in a cofactor-free oxidase.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
4CW6
 
 | | Crystal structure of cofactor-free urate oxidase in complex with the 5-peroxo derivative of 9-metyl uric acid (X-ray dose, 92 kGy) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (5S)-5-(dioxidanyl)-9-methyl-7H-purine-2,6,8-trione, 9-METHYL URIC ACID, ... | | Authors: | Bui, S, Steiner, R.A. | | Deposit date: | 2014-04-01 | | Release date: | 2014-10-29 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Direct evidence for a peroxide intermediate and a reactive enzyme-substrate-dioxygen configuration in a cofactor-free oxidase.
Angew. Chem. Int. Ed. Engl., 53, 2014
|
|
