3F9U
 
 | | Crystal structure of C-terminal domain of putative exported cytochrome C biogenesis-related protein from Bacteroides fragilis | | Descriptor: | 1,2-ETHANEDIOL, NITRATE ION, Putative exported cytochrome C biogenesis-related protein | | Authors: | Chang, C, Tesar, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of C-terminal domain of putative exported cytochrome C biogenesis-related protein from Bacteroides fragilis
To be Published
|
|
3FDI
 
 | | Crystal structure of uncharacterized protein from Eubacterium ventriosum ATCC 27560. | | Descriptor: | CHLORIDE ION, SULFATE ION, uncharacterized protein | | Authors: | Nocek, B, Keigher, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-25 | | Release date: | 2009-01-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of uncharacterized protein from Eubacterium ventriosum ATCC 27560.
To be Published
|
|
3FH0
 
 | | Crystal structure of putative universal stress protein KPN_01444 - ATPase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, putative universal stress protein KPN_01444 | | Authors: | Chang, C, Li, H, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-08 | | Release date: | 2008-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of putative universal stress protein KPN_01444 - ATPase
To be Published
|
|
3FHM
 
 | | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ADENOSINE MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Singer, A.U, Xu, X, Zhang, R, Cui, H, Kudritsdka, M, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the CBS-domain containing protein ATU1752 from Agrobacterium tumefaciens
To be Published
|
|
3EYT
 
 | | Crystal structure of Thioredoxin-like superfamily protein SPOA0173 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, uncharacterized protein SPOA0173 | | Authors: | Chang, C, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-21 | | Release date: | 2008-11-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of Thioredoxin-like superfamily protein SPOA0173
To be Published
|
|
3FDX
 
 | | Putative filament protein / universal stress protein F from Klebsiella pneumoniae. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Osipiuk, J, Volkart, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-26 | | Release date: | 2008-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | X-ray crystal structure of putative filament protein / universal stress protein F from Klebsiella pneumoniae.
To be Published
|
|
5E3E
 
 | | Crystal structure of CdiA-CT/CdiI complex from Y. kristensenii 33638 | | Descriptor: | CdiI immunity protein, Large exoprotein involved in heme utilization or adhesion, SODIUM ION | | Authors: | Michalska, K, Joachimiak, G, Jedrzejczak, R, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-10-02 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The CDI toxin of Yersinia kristensenii is a novel bacterial member of the RNase A superfamily.
Nucleic Acids Res., 45, 2017
|
|
3FXH
 
 | | Crystal structure from the mobile metagenome of Halifax Harbour Sewage Outfall: Integron Cassette Protein HFX_CASS2 | | Descriptor: | Integron gene cassette protein HFX_CASS2 | | Authors: | Sureshan, V, Deshpande, C, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Chang, C, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-20 | | Release date: | 2009-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.837 Å) | | Cite: | Integron gene cassettes: a repository of novel protein folds with distinct interaction sites.
Plos One, 8, 2013
|
|
3FYB
 
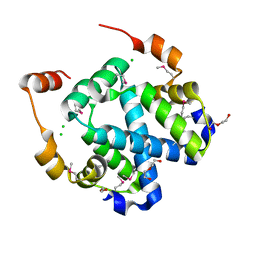 | | Crystal structure of a protein of unknown function (DUF1244) from Alcanivorax borkumensis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-01-22 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a protein of unknown function (DUF1244) from Alcanivorax borkumensis
To be Published
|
|
3G74
 
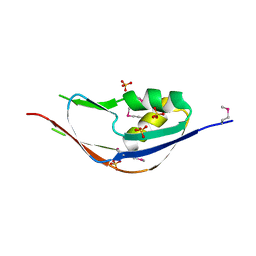 | | Crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560 | | Descriptor: | Protein of unknown function, SULFATE ION | | Authors: | Tan, K, Sather, A, Marshall, N, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The crystal structure of a functionally unknown protein from Eubacterium ventriosum ATCC 27560
To be Published
|
|
3DNP
 
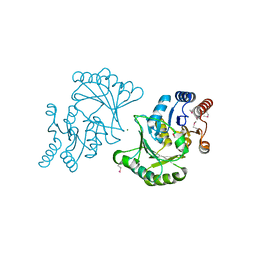 | | Crystal structure of Stress response protein yhaX from Bacillus subtilis | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Stress response protein yhaX | | Authors: | Chang, C, Tesar, C, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-02 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Stress response protein yhaX from Bacillus subtilis
To be Published
|
|
3DTZ
 
 | | Crystal structure of Putative Chlorite dismutase TA0507 | | Descriptor: | FORMIC ACID, Putative Chlorite dismutase TA0507 | | Authors: | Chang, C, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-16 | | Release date: | 2008-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of Putative Chlorite dismutase TA0507
To be Published
|
|
3DDV
 
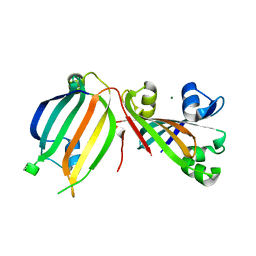 | | The crystal structure of the transcriptional regulator (GntR family) from Enterococcus faecalis V583 | | Descriptor: | MAGNESIUM ION, Transcriptional regulator (GntR family) | | Authors: | Zhang, R, Zhou, M, Bargassa, M, Otwinowski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-06 | | Release date: | 2008-10-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The crystal structure of the transcriptional regulator (GntR family) from Enterococcus faecalis V583
To be Published
|
|
3E0Y
 
 | |
3DT5
 
 | | C_terminal domain of protein of unknown function AF_0924 from Archaeoglobus fulgidus. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Uncharacterized protein AF_0924 | | Authors: | Osipiuk, J, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-14 | | Release date: | 2008-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | X-ray crystal structure of C_terminal domain of protein of unknown function AF_0924 from Archaeoglobus fulgidus.
To be Published
|
|
3DNX
 
 | |
3E0X
 
 | |
3DM8
 
 | | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris | | Descriptor: | DODECYL NONA ETHYLENE GLYCOL ETHER, uncharacterized protein RPA4348 | | Authors: | Cymborowski, M, Chruszcz, M, Skarina, T, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Putative Isomerase from Rhodopseudomonas palustris
To be Published
|
|
3DN7
 
 | | Cyclic nucleotide binding regulatory protein from Cytophaga hutchinsonii. | | Descriptor: | 1,2-ETHANEDIOL, Cyclic nucleotide binding regulatory protein, GLYCEROL | | Authors: | Osipiuk, J, Maltseva, N, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of cyclic nucleotide binding regulatory protein from Cytophaga hutchinsonii.
To be Published
|
|
3DPJ
 
 | | The crystal structure of a TetR transcription regulator from Silicibacter pomeroyi DSS | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Transcription regulator, ... | | Authors: | Tan, K, Li, H, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-08 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a TetR transcription regulator from Silicibacter pomeroyi DSS
To be Published
|
|
3DRW
 
 | | Crystal Structure of a Phosphofructokinase from Pyrococcus horikoshii OT3 with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-specific phosphofructokinase, SODIUM ION | | Authors: | Singer, A.U, Skarina, T, Kochinyan, S, Brown, G, Cuff, M.E, Edwards, A.M, Joachimiak, A, Savchenko, A, Yakunin, A.F, Jia, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-11 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ADP-dependent 6-phosphofructokinase from Pyrococcus horikoshii OT3: structure determination and biochemical characterization of PH1645.
J.Biol.Chem., 284, 2009
|
|
3EEA
 
 | |
3EEF
 
 | | Crystal structure of N-carbamoylsarcosine amidase from thermoplasma acidophilum | | Descriptor: | N-carbamoylsarcosine amidase related protein, ZINC ION | | Authors: | Luo, H.-B, Zheng, H, Chruszcz, M, Zimmerman, M.D, Skarina, T, Egorova, O, Savchenko, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-04 | | Release date: | 2008-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure and molecular modeling study of N-carbamoylsarcosine amidase Ta0454 from Thermoplasma acidophilum.
J.Struct.Biol., 169, 2010
|
|
3E6Q
 
 | | Putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FORMIC ACID, ... | | Authors: | Osipiuk, J, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-08-15 | | Release date: | 2008-08-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | X-ray crystal structure of putative 5-carboxymethyl-2-hydroxymuconate isomerase from Pseudomonas aeruginosa.
To be Published
|
|
3EGL
 
 | | Crystal Structure of DegV Family Protein Cg2579 from Corynebacterium glutamicum | | Descriptor: | DegV family protein, FORMIC ACID, PALMITIC ACID | | Authors: | Kim, Y, Tesar, C, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-11 | | Release date: | 2008-09-23 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Crystal Structure of DegV Family Protein Cg2579 from Corynebacterium glutamicum
To be Published
|
|
