6DGB
 
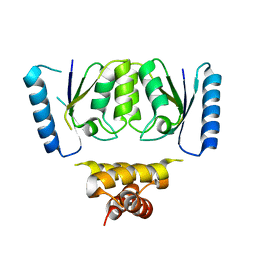 | | Crystal structure of the C-terminal catalytic domain of IS1535 TnpA, an IS607-like serine recombinase | | Descriptor: | IS607 family transposase IS1535 | | Authors: | Chen, W.Y, Hancock, S.P, Cascio, D, Johnson, R.C. | | Deposit date: | 2018-05-17 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Multiple serine transposase dimers assemble the transposon-end synaptic complex during IS607-family transposition.
Elife, 7, 2018
|
|
3B6S
 
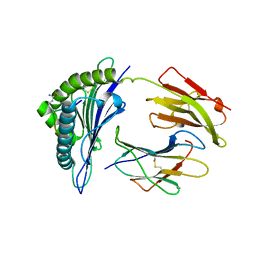 | | Crystal Structure of hla-b*2705 Complexed with the Citrullinated Vasoactive Intestinal Peptide Type 1 Receptor (vipr) Peptide (residues 400-408) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | Authors: | Beltrami, A, Rossmann, M, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Citrullination-dependent Differential Presentation of a Self-peptide by HLA-B27 Subtypes.
J.Biol.Chem., 283, 2008
|
|
7MKV
 
 | |
5VND
 
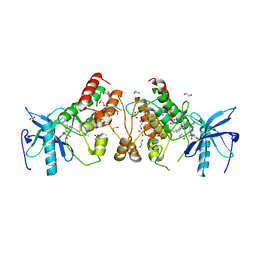 | | Crystal structure of FGFR1-Y563C (FGFR4 surrogate) covalently bound to H3B-6527 | | Descriptor: | 1,2-ETHANEDIOL, Fibroblast growth factor receptor 1, N-{2-[(6-{[(2,6-dichloro-3,5-dimethoxyphenyl)carbamoyl](methyl)amino}pyrimidin-4-yl)amino]-5-(4-ethylpiperazin-1-yl)phenyl}propanamide, ... | | Authors: | Tsai, J.H.C, Reynolds, D, Fekkes, P, Smith, P, Larsen, N.A. | | Deposit date: | 2017-04-30 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | H3B-6527 Is a Potent and Selective Inhibitor of FGFR4 in FGF19-Driven Hepatocellular Carcinoma.
Cancer Res., 77, 2017
|
|
8UR9
 
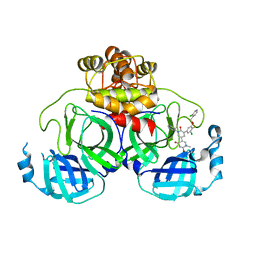 | | Crystal Structure of the SARS-CoV-2 Main Protease in Complex with Compound 61 | | Descriptor: | (5P)-5-[(1P,3M,3'P)-3-{3-chloro-5-[(2-chlorophenyl)methoxy]-4-fluorophenyl}-2-oxo-2H-[1,3'-bipyridin]-5-yl]-1-methylpyrimidine-2,4(1H,3H)-dione, 3C-like proteinase nsp5 | | Authors: | Papini, C, Zhang, C.H, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-10-25 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proof-of-concept studies with a computationally designed M pro inhibitor as a synergistic combination regimen alternative to Paxlovid.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VBB
 
 | |
8VBA
 
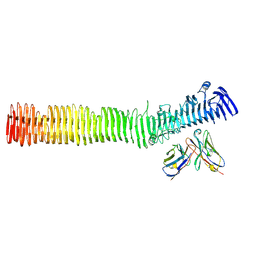 | |
5DCC
 
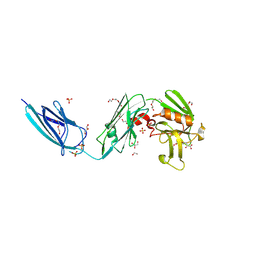 | | X-RAY CRYSTAL STRUCTURE OF a TEBIPENEM ADDUCT OF L,D TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | (4S)-4-methyl-2,5,7-trioxoheptanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pan, Y, Basta, L, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-23 | | Release date: | 2016-09-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
5DC2
 
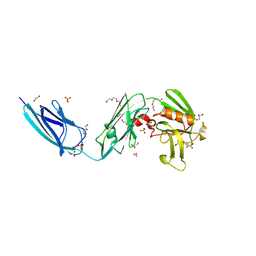 | | X-RAY CRYSTAL STRUCTURE OF A ENZYMATICALLY DEGRADED BIAPENEM-ADDUCT OF L,D-TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | (4S)-4-methyl-2,5,7-trioxoheptanoic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pan, Y, Basta, L, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-23 | | Release date: | 2016-10-05 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.182 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
5D7H
 
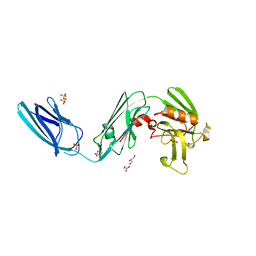 | | X-RAY CRYSTAL STRUCTURE OF L,D TRANSPEPTIDASE 2 FROM MYCOBACTERIUM TUBERCULOSIS | | Descriptor: | GLYCEROL, L,D-transpeptidase 2, SULFATE ION | | Authors: | Saavedra, H, Basta, L.A, Lamichhane, G, Bianchet, M.A. | | Deposit date: | 2015-08-13 | | Release date: | 2016-09-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural insight into the inactivation of Mycobacterium tuberculosis non-classical transpeptidase LdtMt2 by biapenem and tebipenem.
BMC Biochem., 18, 2017
|
|
2PUJ
 
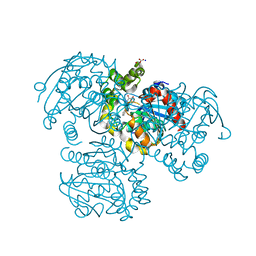 | | Crystal Structure of the S112A/H265A double mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, in complex with its substrate HOPDA | | Descriptor: | (2E,4E)-2-HYDROXY-6-OXO-6-PHENYLHEXA-2,4-DIENOIC ACID, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION, ... | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | The Tautomeric Half-reaction of BphD, a C-C Bond Hydrolase: KINETIC AND STRUCTURAL EVIDENCE SUPPORTING A KEY ROLE FOR HISTIDINE 265 OF THE CATALYTIC TRIAD.
J.Biol.Chem., 282, 2007
|
|
2PU7
 
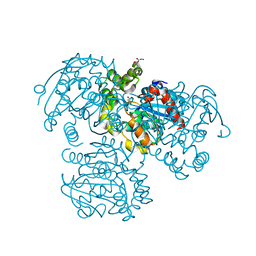 | | Crystal Structure of S112A/H265A double mutant of a C-C hydrolase, BphD, from Burkholderia xenovorans LB400 | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION, SODIUM ION | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The Tautomeric Half-reaction of BphD, a C-C Bond Hydrolase: KINETIC AND STRUCTURAL EVIDENCE SUPPORTING A KEY ROLE FOR HISTIDINE 265 OF THE CATALYTIC TRIAD.
J.Biol.Chem., 282, 2007
|
|
2PUH
 
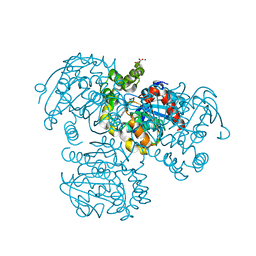 | | Crystal Structure of the S112A mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, in complex with its substrate HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION, ... | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-05-09 | | Release date: | 2007-05-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Tautomeric Half-reaction of BphD, a C-C Bond Hydrolase: KINETIC AND STRUCTURAL EVIDENCE SUPPORTING A KEY ROLE FOR HISTIDINE 265 OF THE CATALYTIC TRIAD.
J.Biol.Chem., 282, 2007
|
|
2PU5
 
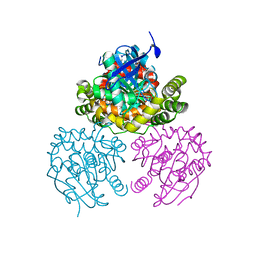 | | Crystal Structure of a C-C bond hydrolase, BphD, from Burkholderia xenovorans LB400 | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-05-08 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Tautomeric Half-reaction of BphD, a C-C Bond Hydrolase: KINETIC AND STRUCTURAL EVIDENCE SUPPORTING A KEY ROLE FOR HISTIDINE 265 OF THE CATALYTIC TRIAD.
J.Biol.Chem., 282, 2007
|
|
2RI6
 
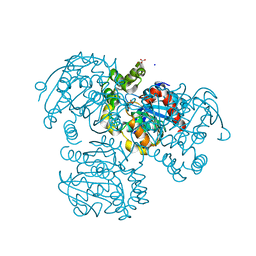 | | Crystal Structure of S112A mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400 | | Descriptor: | 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, MALONATE ION, SODIUM ION | | Authors: | Bhowmik, S, Bolin, J.T. | | Deposit date: | 2007-10-10 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The tautomeric half-reaction of BphD, a C-C bond hydrolase. Kinetic and structural evidence supporting a key role for histidine 265 of the catalytic triad.
J.Biol.Chem., 282, 2007
|
|
6BOI
 
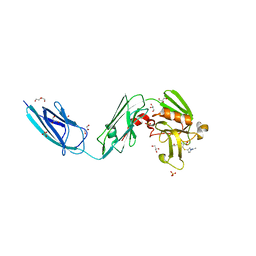 | | Crystal Structure of LdtMt2 (56-408) with a panipenem adduct at the active site cysteine-354 | | Descriptor: | (3S,5R)-5-[(2R,3R)-1,3-dihydroxybutan-2-yl]-3-({(3R)-1-[(1E)-ethanimidoyl]pyrrolidin-3-yl}sulfanyl)-L-proline, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Saavedra, H, Bianchet, M.A. | | Deposit date: | 2017-11-20 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structures and Mechanism of Inhibition of Mycobacterium tuberculosis L,D-transpeptidase 2 by Panipenem
To Be Published
|
|
2NBY
 
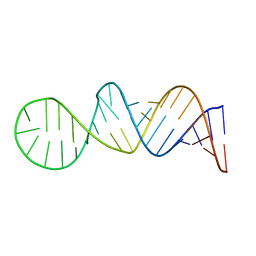 | |
2NBX
 
 | |
2NBZ
 
 | |
2NC0
 
 | |
2NC1
 
 | |
4PPG
 
 | | Mycobacterium tuberculosis RecA citrate bound low temperature structure IIA-BR | | Descriptor: | CITRATE ANION, Protein RecA, 1st part, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PPQ
 
 | | Mycobacterium tuberculosis RecA citrate bound low temperature structure IIA-CR | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-02-27 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PQY
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-N4 | | Descriptor: | GLYCEROL, Protein RecA, 1st part, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-05 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
4PTL
 
 | | Mycobacterium tuberculosis RecA glycerol bound low temperature structure IIC-GM | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein RecA, ... | | Authors: | Chandran, A.V, Prabu, J.R, Patil, N.K, Muniyappa, K, Vijayan, M. | | Deposit date: | 2014-03-11 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on Mycobacterium tuberculosis RecA: Molecular plasticity and interspecies variability
J.Biosci., 40, 2015
|
|
