2B4T
 
 | | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 Angstrom resolution reveals intriguing extra electron density in the active site | | Descriptor: | 4-(2-AMINOETHYL)BENZENESULFONYL FLUORIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Robien, M.A, Bosch, J, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-09-26 | | Release date: | 2005-10-04 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of glyceraldehyde-3-phosphate dehydrogenase from Plasmodium falciparum at 2.25 A resolution reveals intriguing extra electron density in the active site
Proteins, 62, 2006
|
|
2B9S
 
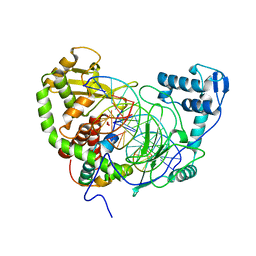 | | Crystal Structure of heterodimeric L. donovani topoisomerase I-vanadate-DNA complex | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*T)-3', 5'-D(*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3', 5'-D(*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3', ... | | Authors: | Davies, D.R, Hol, W.G.J. | | Deposit date: | 2005-10-12 | | Release date: | 2006-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | The Structure of the Transition State of the Heterodimeric Topoisomerase I of Leishmania donovani as a Vanadate Complex with Nicked DNA.
J.Mol.Biol., 357, 2006
|
|
2EPH
 
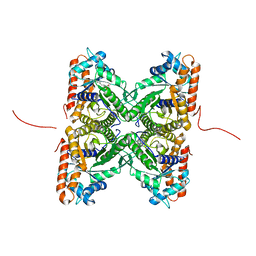 | | Crystal structure of fructose-bisphosphate aldolase from Plasmodium falciparum in complex with TRAP-tail determined at 2.7 angstrom resolution | | Descriptor: | Fructose-bisphosphate aldolase, PbTRAP | | Authors: | Bosch, J, Buscaglia, C.A, Krumm, B, Cardozo, T, Nussenzweig, V, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2007-03-30 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aldolase provides an unusual binding site for thrombospondin-related anonymous protein in the invasion machinery of the malaria parasite.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1LTA
 
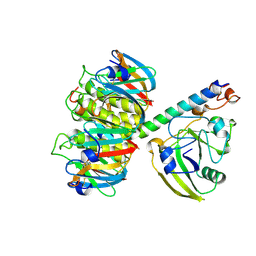 | | 2.2 ANGSTROMS CRYSTAL STRUCTURE OF E. COLI HEAT-LABILE ENTEROTOXIN (LT) WITH BOUND GALACTOSE | | Descriptor: | HEAT-LABILE ENTEROTOXIN, SUBUNIT A, SUBUNIT B, ... | | Authors: | Merritt, E.A, Sixma, T.K, Kalk, K.H, Van Zanten, B.A.M, Hol, W.G.J. | | Deposit date: | 1993-09-15 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Galactose-binding site in Escherichia coli heat-labile enterotoxin (LT) and cholera toxin (CT).
Mol.Microbiol., 13, 1994
|
|
1AG1
 
 | |
1A36
 
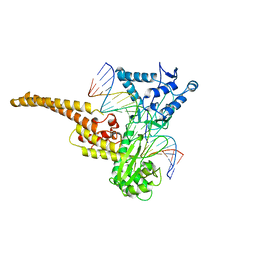 | | TOPOISOMERASE I/DNA COMPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*TP*TP*AP*GP*AP*AP*AP*AP*A P*TP*TP*TP*TP*T)- 3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*AP*AP*GP*TP*C P*TP*TP*TP*TP*T)- 3'), TOPOISOMERASE I | | Authors: | Stewart, L, Redinbo, M.R, Qiu, X, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 1998-01-29 | | Release date: | 1998-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A model for the mechanism of human topoisomerase I.
Science, 279, 1998
|
|
1BP2
 
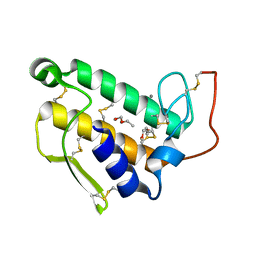 | | STRUCTURE OF BOVINE PANCREATIC PHOSPHOLIPASE A2 AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PHOSPHOLIPASE A2 | | Authors: | Dijkstra, B.W, Kalk, K.H, Hol, W.G.J, Drenth, J. | | Deposit date: | 1981-04-06 | | Release date: | 1981-05-21 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of bovine pancreatic phospholipase A2 at 1.7A resolution.
J.Mol.Biol., 147, 1981
|
|
13PK
 
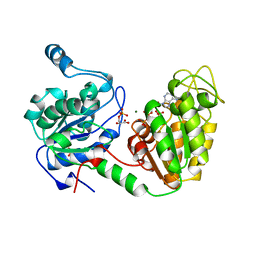 | | TERNARY COMPLEX OF PHOSPHOGLYCERATE KINASE FROM TRYPANOSOMA BRUCEI | | Descriptor: | 3-PHOSPHOGLYCERATE KINASE, 3-PHOSPHOGLYCERIC ACID, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bernstein, B.E, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 1996-11-23 | | Release date: | 1997-12-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Synergistic effects of substrate-induced conformational changes in phosphoglycerate kinase activation.
Nature, 385, 1997
|
|
1A31
 
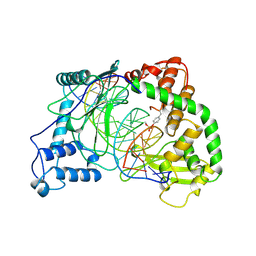 | | HUMAN RECONSTITUTED DNA TOPOISOMERASE I IN COVALENT COMPLEX WITH A 22 BASE PAIR DNA DUPLEX | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*GP*AP*CP*5IUP*5IU*TP*GP*AP*AP*AP*AP*AP*5IUP*5IUP*5IUP*5IUP*T)-3'), DNA (5'-D(*AP*AP*AP*AP*AP*TP*5IUP*5IUP*5IUP*5IUP*CP*AP*AP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), PROTEIN (TOPOISOMERASE I) | | Authors: | Redinbo, M.R, Stewart, L, Kuhn, P, Champoux, J.J, Hol, W.G.J. | | Deposit date: | 1998-01-27 | | Release date: | 1998-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA.
Science, 279, 1998
|
|
1A5C
 
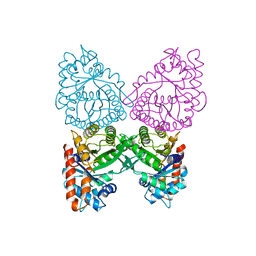 | | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE FROM PLASMODIUM FALCIPARUM | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE | | Authors: | Kim, H, Certa, U, Dobeli, H, Jakob, P, Hol, W.G.J. | | Deposit date: | 1998-02-13 | | Release date: | 1998-06-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of fructose-1,6-bisphosphate aldolase from the human malaria parasite Plasmodium falciparum.
Biochemistry, 37, 1998
|
|
16PK
 
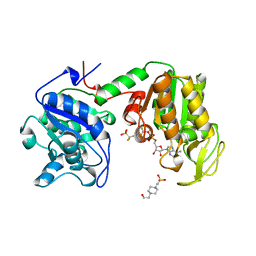 | | PHOSPHOGLYCERATE KINASE FROM TRYPANOSOMA BRUCEI BISUBSTRATE ANALOG | | Descriptor: | 1,1,5,5-TETRAFLUOROPHOSPHOPENTYLPHOSPHONIC ACID ADENYLATE ESTER, 3-PHOSPHOGLYCERATE KINASE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID | | Authors: | Bernstein, B.E, Bressi, J, Blackburn, M, Gelb, M, Hol, W.G.J. | | Deposit date: | 1998-05-18 | | Release date: | 1998-11-25 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A bisubstrate analog induces unexpected conformational changes in phosphoglycerate kinase from Trypanosoma brucei.
J.Mol.Biol., 279, 1998
|
|
1HTL
 
 | |
1LLA
 
 | |
1LTT
 
 | |
1LTG
 
 | |
1LTB
 
 | |
1DG5
 
 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND TRIMETHOPRIM | | Descriptor: | DIHYDROFOLATE REDUCTASE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-23 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
1DG8
 
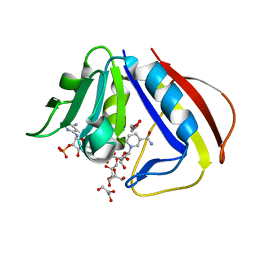 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH | | Descriptor: | DIHYDROFOLATE REDUCTASE, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-23 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
1DF7
 
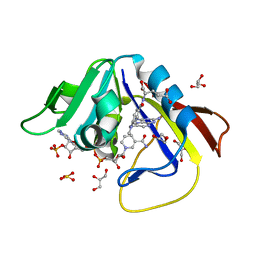 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND METHOTREXATE | | Descriptor: | DIHYDROFOLATE REDUCTASE, GLYCEROL, METHOTREXATE, ... | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-17 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
1DG7
 
 | | DIHYDROFOLATE REDUCTASE OF MYCOBACTERIUM TUBERCULOSIS COMPLEXED WITH NADPH AND 4-BROMO WR99210 | | Descriptor: | 1-[3-(4-BROMO-PHENOXY)-PROPOXY]-6,6-DIMETHYL-1.6-DIHYDRO-[1,3,5]TRIAZINE-2,4-DIAMINE, DIHYDROFOLATE REDUCTASE, GLYCEROL, ... | | Authors: | Li, R, Sirawaraporn, R, Chitnumsub, P, Sirawaraporn, W, Wooden, J, Athappilly, F, Turley, S, Hol, W.G. | | Deposit date: | 1999-11-23 | | Release date: | 2000-03-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of M. tuberculosis dihydrofolate reductase reveals opportunities for the design of novel tuberculosis drugs.
J.Mol.Biol., 295, 2000
|
|
1NHP
 
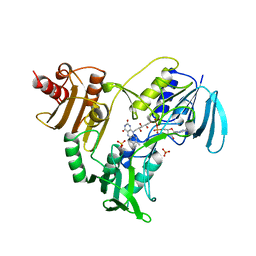 | | CRYSTALLOGRAPHIC ANALYSES OF NADH PEROXIDASE CYS42ALA AND CYS42SER MUTANTS: ACTIVE SITE STRUCTURE, MECHANISTIC IMPLICATIONS, AND AN UNUSUAL ENVIRONMENT OF ARG303 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE, SULFATE ION | | Authors: | Mande, S.S, Claiborne, A, Hol, W.G.J. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic analyses of NADH peroxidase Cys42Ala and Cys42Ser mutants: active site structures, mechanistic implications, and an unusual environment of Arg 303.
Biochemistry, 34, 1995
|
|
1E4Q
 
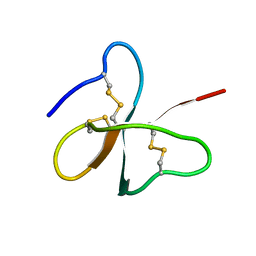 | | Solution structure of the human defensin hBD-2 | | Descriptor: | BETA-DEFENSIN 2 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
1E4T
 
 | | Solution structure of the mouse defensin mBD-7 | | Descriptor: | Beta-defensin 7 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2018-06-20 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human and murine beta-defensins reveals structural conservation in the absence of significant sequence similarity.
Protein Sci., 10, 2001
|
|
1EJ9
 
 | | CRYSTAL STRUCTURE OF HUMAN TOPOISOMERASE I DNA COMPLEX | | Descriptor: | DNA (5'-D(*C*AP*AP*AP*AP*AP*GP*AP*CP*TP*CP*AP*GP*AP*AP*AP*AP*AP*TP*TP*TP*TP*T)-3'), DNA (5'-D(*C*AP*AP*AP*AP*AP*TP*TP*TP*TP*TP*CP*TP*GP*AP*GP*TP*CP*TP*TP*TP*TP*T)-3'), DNA TOPOISOMERASE I | | Authors: | Redinbo, M.R, Champoux, J.J, Hol, W.G. | | Deposit date: | 2000-03-01 | | Release date: | 2000-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA.
Biochemistry, 39, 2000
|
|
1E4S
 
 | | Solution structure of the human defensin hBD-1 | | Descriptor: | BETA-DEFENSIN 1 | | Authors: | Bauer, F, Schweimer, K, Kluver, E, Adermann, K, Forssmann, W.G, Roesch, P, Sticht, H. | | Deposit date: | 2000-07-12 | | Release date: | 2001-07-12 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of Human and Murine Beta-Defensins Reveals Structural Conservation in the Absence of Significant Sequence Similarity
Protein Sci., 10, 2001
|
|
