4O1Q
 
 | |
4Y5R
 
 | | Crystal Structure of a T67A MauG/pre-Methylamine Dehydrogenase Complex | | Descriptor: | CALCIUM ION, HEME C, Methylamine dehydrogenase heavy chain, ... | | Authors: | Li, C, Wilmot, C.M. | | Deposit date: | 2015-02-11 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A T67A mutation in the proximal pocket of the high-spin heme of MauG stabilizes formation of a mixed-valent Fe(II)/Fe(III) state and enhances charge resonance stabilization of the bis-Fe(IV) state.
Biochim.Biophys.Acta, 1847, 2015
|
|
2LN4
 
 | |
2MBC
 
 | | Solution Structure of human holo-PRL-3 in complex with vanadate | | Descriptor: | Protein tyrosine phosphatase type IVA 3 | | Authors: | Jeong, K, Kang, D, Kim, J, Shin, S, Jin, B, Lee, C, Kim, E, Jeon, Y.H, Kim, Y. | | Deposit date: | 2013-07-29 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and backbone dynamics of vanadate-bound PRL-3: comparison of 15N nuclear magnetic resonance relaxation profiles of free and vanadate-bound PRL-3.
Biochemistry, 53, 2014
|
|
2POO
 
 | | THERMOSTABLE PHYTASE IN FULLY CALCIUM LOADED STATE | | Descriptor: | CALCIUM ION, PROTEIN (PHYTASE) | | Authors: | Ha, N.-C, Oh, B.-H. | | Deposit date: | 1999-04-16 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
3QID
 
 | | Crystal structures and functional analysis of murine norovirus RNA-dependent RNA polymerase | | Descriptor: | GLYCEROL, MANGANESE (III) ION, RNA dependent RNA polymerase, ... | | Authors: | Kim, K.H, Intekhab, A, Lee, J.H. | | Deposit date: | 2011-01-27 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase.
J.Gen.Virol., 92, 2011
|
|
5KTG
 
 | |
2X29
 
 | | Crystal structure of human4-1BB ligand ectodomain | | Descriptor: | TUMOR NECROSIS FACTOR LIGAND SUPERFAMILY MEMBER 9 | | Authors: | Won, E.Y, Cho, H.S. | | Deposit date: | 2010-01-12 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of the Trimer of Human 4-1Bb Ligand is Unique Among Members of the Tumor Necrosis Factor Superfamily.
J.Biol.Chem., 285, 2010
|
|
5AI1
 
 | | Crystal structure of ketosteroid isomerase containing Y32F, D40N, Y57F and Y119F mutations in the equilenin-bound form | | Descriptor: | EQUILENIN, KETOSTEROID ISOMERASE | | Authors: | Cha, H.J, Jeong, J.H, Kim, Y.G. | | Deposit date: | 2015-02-11 | | Release date: | 2015-05-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Contribution of a Low-Barrier Hydrogen Bond to Catalysis is not Significant in Ketosteroid Isomerase.
Mol.Cells, 38, 2015
|
|
2HI3
 
 | | Solution structure of the homeodomain-only protein HOP | | Descriptor: | Homeodomain-only protein | | Authors: | Mackay, J.P, Kook, H, Epstein, J.A, Simpson, R.J, Yung, W.W. | | Deposit date: | 2006-06-28 | | Release date: | 2007-01-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Analysis of the structure and function of the transcriptional coregulator HOP
Biochemistry, 45, 2006
|
|
4L3G
 
 | |
4L3H
 
 | |
1POO
 
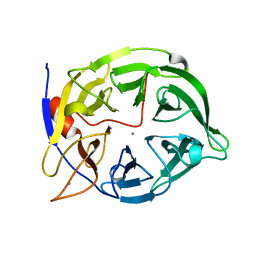 | | THERMOSTABLE PHYTASE FROM BACILLUS SP | | Descriptor: | CALCIUM ION, PROTEIN (PHYTASE) | | Authors: | Oh, B.H, Ha, N.C. | | Deposit date: | 1999-04-16 | | Release date: | 2000-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states.
Nat.Struct.Biol., 7, 2000
|
|
4L1Q
 
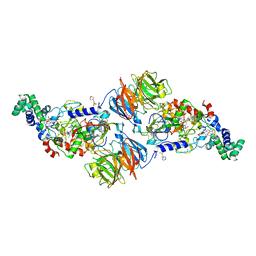 | |
2LA2
 
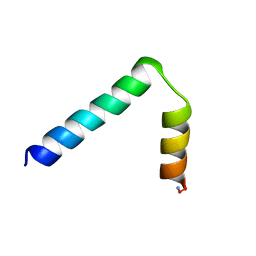 | |
8GUW
 
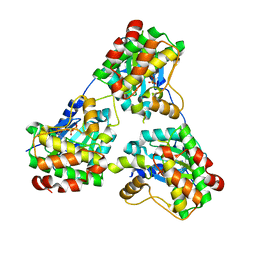 | |
4FA5
 
 | |
4FB1
 
 | |
4FAV
 
 | |
4FAN
 
 | |
4FA1
 
 | |
4FA9
 
 | |
4FA4
 
 | |
8HAL
 
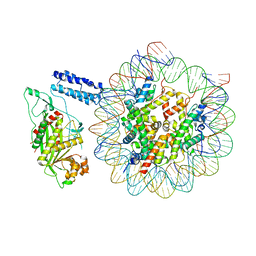 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 1 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAG
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (3.2 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
