6ZB5
 
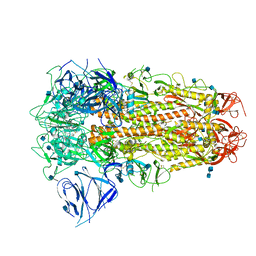 | | SARS CoV-2 Spike protein, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-07 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
5K0Z
 
 | | Cryo-EM structure of lactate dehydrogenase (LDH) in inhibitor-bound state | | Descriptor: | L-lactate dehydrogenase B chain | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
6ZB4
 
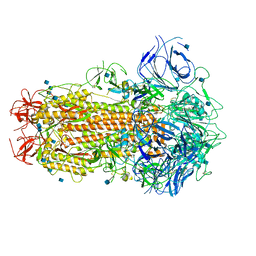 | | SARS CoV-2 Spike protein, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Burucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Science, 370, 2020
|
|
5WDR
 
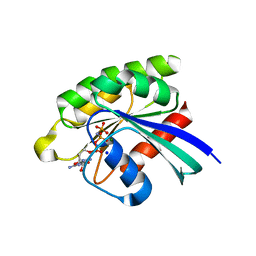 | | Choanoflagellate Salpingoeca rosetta Ras with GMP-PNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras protein, ... | | Authors: | Kondo, Y, Gee, C.L, Kuriyan, J. | | Deposit date: | 2017-07-05 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Deconstruction of the Ras switching cycle through saturation mutagenesis.
Elife, 6, 2017
|
|
6ARO
 
 | |
5K11
 
 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) in inhibitor-bound state | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
5K10
 
 | | Cryo-EM structure of isocitrate dehydrogenase (IDH1) | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Merk, A, Bartesaghi, A, Banerjee, S, Falconieri, V, Rao, P, Earl, L, Milne, J, Subramaniam, S. | | Deposit date: | 2016-05-17 | | Release date: | 2016-06-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery.
Cell, 165, 2016
|
|
3TSD
 
 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames complexed with XMP | | Descriptor: | D(-)-TARTARIC ACID, Inosine-5'-monophosphate dehydrogenase, SULFATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
3TSB
 
 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames | | Descriptor: | Inosine-5'-monophosphate dehydrogenase, PHOSPHATE ION | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-12 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
6D0X
 
 | | Crystal structure of chimeric H.2140 / K.1210 Fab in complex with circumsporozoite protein NANP3 | | Descriptor: | 1210 Antibody, light chain, 2140 Antibody, ... | | Authors: | Scally, S.W, Bosch, A, Imkeller, K, Wardemann, H, Julien, J.P. | | Deposit date: | 2018-04-11 | | Release date: | 2018-06-13 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Antihomotypic affinity maturation improves human B cell responses against a repetitive epitope.
Science, 360, 2018
|
|
6D01
 
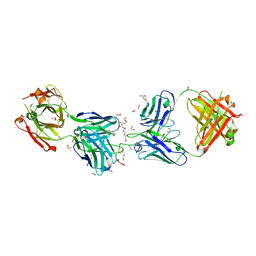 | | Crystal structure of 1210 Fab in complex with circumsporozoite protein NANP5 | | Descriptor: | 1,2-ETHANEDIOL, 1210 Antibody, Light chain, ... | | Authors: | Scally, S.W, Bosch, A, Imkeller, K, Wardemann, H, Julien, J.P. | | Deposit date: | 2018-04-09 | | Release date: | 2018-06-13 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Antihomotypic affinity maturation improves human B cell responses against a repetitive epitope.
Science, 360, 2018
|
|
6D11
 
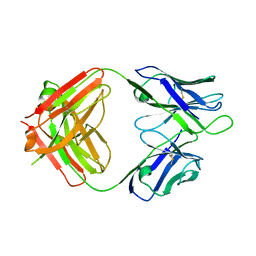 | | Crystal structure of 1450 Fab in complex with circumsporozoite protein NANP5 | | Descriptor: | 1450 Antibody, Heavy chain, Light chain, ... | | Authors: | Scally, S.W, Bosch, A, Imkeller, K, Wardemann, H, Julien, J.P. | | Deposit date: | 2018-04-11 | | Release date: | 2018-06-13 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Antihomotypic affinity maturation improves human B cell responses against a repetitive epitope.
Science, 360, 2018
|
|
1MLU
 
 | |
2OKY
 
 | |
6EH7
 
 | |
6EH6
 
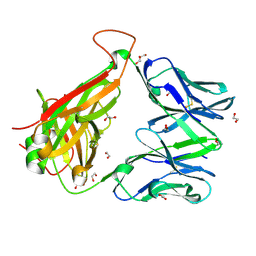 | |
6EH5
 
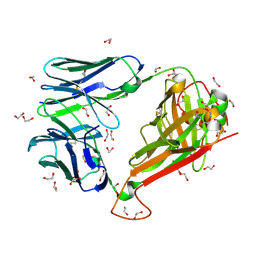 | |
5BK5
 
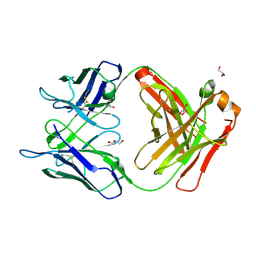 | | Crystal structure of the anti-circumsporozoite protein 663 germline antibody | | Descriptor: | 663 germline antibody, heavy chain, light chain, ... | | Authors: | Scally, S.W, Bosch, A, Triller, G, Wardemann, H, Julien, J.P. | | Deposit date: | 2017-09-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Natural Parasite Exposure Induces Protective Human Anti-Malarial Antibodies.
Immunity, 47, 2017
|
|
5BK0
 
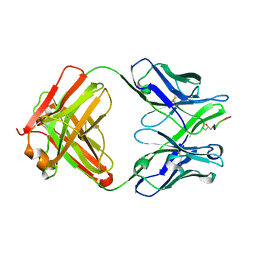 | | Crystal structure of 663 Fab bound to circumsporozoite protein NANP 5-mer | | Descriptor: | 663 Antibody, heavy chain, light chain, ... | | Authors: | Scally, S.W, Bosch, A, Triller, G, Wardemann, H, Julien, J.P. | | Deposit date: | 2017-09-12 | | Release date: | 2017-12-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Natural Parasite Exposure Induces Protective Human Anti-Malarial Antibodies.
Immunity, 47, 2017
|
|
6EH8
 
 | |
6EH9
 
 | |
4D9Q
 
 | |
2C1E
 
 | | Crystal structures of caspase-3 in complex with aza-peptide Michael acceptor inhibitors. | | Descriptor: | AZA-PEPTIDE INHIBITOR (5S, 8R, 11S)-8-(2-CARBOXYETHYL)-5-(CARBOXYMETHYL)-14-(4-ETHOXY-4-OXOBUTANOYL)-11-(1-METHYLETHYL)-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,13,14-PENTAAZAHEXADECAN -16-OIC ACID, ... | | Authors: | Grutter, M.G. | | Deposit date: | 2005-09-14 | | Release date: | 2006-09-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Design, Synthesis, and Evaluation of Aza-Peptide Michael Acceptors as Selective and Potent Inhibitors of Caspases-2, -3, -6, -7, -8, -9, and - 10.
J.Med.Chem., 49, 2006
|
|
6FR3
 
 | | 003 TCR Study of CDR Loop Flexibility | | Descriptor: | 003 TCR Alpha Chain, 003 TCR Beta Chain, 1,2-ETHANEDIOL, ... | | Authors: | Rizkallah, P.J, Cole, D.K. | | Deposit date: | 2018-02-15 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | In Silicoand Structural Analyses Demonstrate That Intrinsic Protein Motions Guide T Cell Receptor Complementarity Determining Region Loop Flexibility.
Front Immunol, 9, 2018
|
|
6FRC
 
 | |
