5I04
 
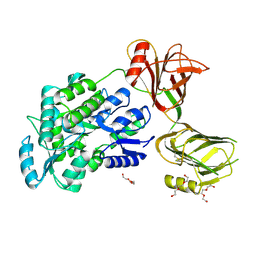 | | Crystal structure of the orphan region of human endoglin/CD105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Endoglin, TRIETHYLENE GLYCOL, ... | | Authors: | Saito, T, Bokhove, M, de Sanctis, D, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
5NNE
 
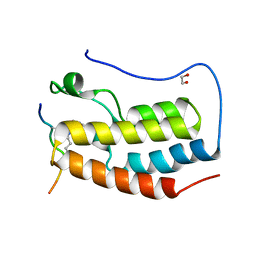 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated TOP2A peptide (K1201ac/K1204ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, GKA(ALY)GK(ALY)TQMY | | Authors: | Filippakopoulos, P, Picaud, S, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
5NND
 
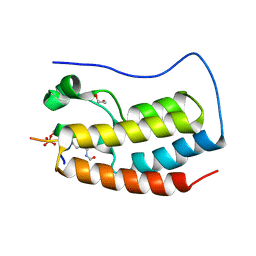 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a diacetylated histone 4 peptide (H3K9ac/K14ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Histone H3 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
4E6P
 
 | |
4HKT
 
 | |
3HRW
 
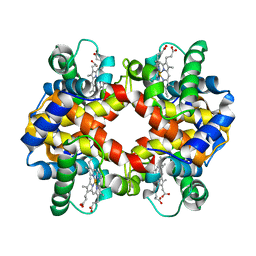 | | Crystal structure of hemoglobin from mouse (Mus musculus)at 2.8 | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta-1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sundaresan, S.S, Ramesh, P, Ponnuswamy, M.N. | | Deposit date: | 2009-06-10 | | Release date: | 2009-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of hemoglobin from mouse (Mus musculus) compared with those from other small animals and humans.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4HC8
 
 | |
4JWT
 
 | |
4JOS
 
 | | Crystal structure of a putative 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Francisella philomiragia ATCC 25017 (Target NYSGRC-029335) | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Adenosylhomocysteine nucleosidase, ... | | Authors: | Sampathkumar, P, Schramm, V.L, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-03-18 | | Release date: | 2013-04-03 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a putative 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Francisella philomiragia ATCC 25017 (Target NYSGRC-029335)
to be published
|
|
4MJV
 
 | | Influenza Neuraminidase in complex with a novel antiviral compound | | Descriptor: | (2E,5S,9R,10S)-10-(acetylamino)-2-imino-4-oxo-9-(pentan-3-yloxy)-1-thia-3-azaspiro[4.5]dec-6-ene-7-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S. | | Deposit date: | 2013-09-04 | | Release date: | 2014-01-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Serendipitous discovery of a potent influenza virus a neuraminidase inhibitor.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MJU
 
 | | Influenza Neuraminidase in complex with a novel antiviral compound | | Descriptor: | (5R,9R,10S)-10-(acetylamino)-2-amino-4-oxo-9-(pentan-3-yloxy)-1-thia-3-azaspiro[4.5]deca-2,6-diene-7-carboxylic acid, CALCIUM ION, Neuraminidase | | Authors: | Kerry, P.S. | | Deposit date: | 2013-09-04 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Serendipitous discovery of a potent influenza virus a neuraminidase inhibitor.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3W09
 
 | | Influenza virus neuraminidase subtype N9 (TERN) complexed with 2,3-dif guanidino-neu5ac2en inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-acetamido-2,6-anhydro-4-carbamimidamido-3,4,5-trideoxy-3-fluoro-D-glycero-D-galacto-non-2-enonic acid, ... | | Authors: | Streltsov, V.A. | | Deposit date: | 2012-10-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism-based covalent neuraminidase inhibitors with broad-spectrum influenza antiviral activity
Science, 340, 2013
|
|
4G8S
 
 | | Crystal Structure Of a Putative Nitroreductase from Geobacter sulfurreducens PCA (Target PSI-013445) | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Nitroreductase family protein | | Authors: | Kumar, P.R, Bhosle, R, Hillerich, B, Seidel, R, Toro, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a nitroreductase from Geobacter sulfurreducens PCA
to be published
|
|
3FS4
 
 | | Crystal structure determination of Ostrich hemoglobin at 2.2 Angstrom resolution | | Descriptor: | Hemoglobin subunit alpha-A, Hemoglobin subunit beta, OXYGEN MOLECULE, ... | | Authors: | Sundaresan, S.S, Ramesh, P, Sivakumar, K, Ponnuswamy, M.N. | | Deposit date: | 2009-01-09 | | Release date: | 2009-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural studies of hemoglobin from two flightless birds, ostrich and turkey: insights into their differing oxygen-binding properties.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
4GI5
 
 | | Crystal Structure Of a Putative quinone reductase from Klebsiella pneumoniae (Target PSI-013613) | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a quinone reductase from Klebsiella pneumoniae with bound FAD
to be published
|
|
4KS1
 
 | | Influenza neuraminidase in complex with antiviral compound (3S,4R,5R)-4-(acetylamino)-3-amino-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid | | Descriptor: | (3S,4R,5R)-4-(acetylamino)-3-amino-5-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Kerry, P.S, Russell, R.J.M. | | Deposit date: | 2013-05-17 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for a class of nanomolar influenza A neuraminidase inhibitors.
Sci Rep, 3, 2013
|
|
4GIC
 
 | | Crystal Structure Of a Putative Histidinol dehydrogenase (Target PSI-014034) from Methylococcus capsulatus | | Descriptor: | Histidinol dehydrogenase, SULFATE ION | | Authors: | Kumar, P.R, Ahmed, M, Banu, N, Bhosle, R, Bonanno, J, Chamala, S, Chowdhury, S, Gizzi, A, Glen, S, Hammonds, J, Hillerich, B, Love, J.D, Seidel, R, Stead, M, Toro, R, Washington, E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-08 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of a putative Histidinol dehydrogenase from Methylococcus capsulatus
to be published
|
|
