7EA8
 
 | |
7EA5
 
 | |
7E5R
 
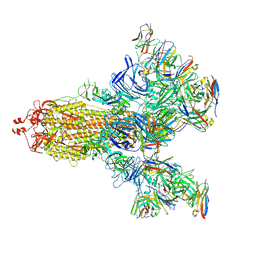 | | SARS-CoV-2 S trimer with three-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
7E8O
 
 | |
7E8K
 
 | |
7E8J
 
 | |
7DZE
 
 | | Fabp ground state captured by XFELs | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZF
 
 | | Intermediate of FABP with a delay time of 10 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZG
 
 | | Intermediate of FABP with a delay time of 30 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZH
 
 | | intermediate of FABP with a delay time of 100 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZI
 
 | | intermediate of FABP with a delay time of 300 ns | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZJ
 
 | | Fabp protein before hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZK
 
 | | Fabp protein after hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZL
 
 | | A69C-M71L mutant of Fabp protein | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7EXD
 
 | | Lasmiditan-bound serotonin 1F (5-HT1F) receptor-Gi protein complex | | Descriptor: | 2,4,6-tris(fluoranyl)-N-[6-(1-methylpiperidin-4-yl)carbonylpyridin-2-yl]benzamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S, Xu, P, Jiang, Y, Xu, H.E. | | Deposit date: | 2021-05-27 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for recognition of anti-migraine drug lasmiditan by the serotonin receptor 5-HT 1F -G protein complex.
Cell Res., 31, 2021
|
|
7FIA
 
 | | Structure of AcrIF23 | | Descriptor: | AcrIF23 | | Authors: | Ren, J, Yue, F. | | Deposit date: | 2021-07-30 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural and mechanistic insights into the inhibition of type I-F CRISPR-Cas system by anti-CRISPR protein AcrIF23.
J.Biol.Chem., 298, 2022
|
|
7XP1
 
 | | Crystal structure of PmiR from Pseudomonas aeruginosa | | Descriptor: | ALPHA-METHYLISOCITRIC ACID, GLYCEROL, Probable transcriptional regulator, ... | | Authors: | Zhang, Y.X, Liang, H.H, Gan, J.H. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | PmiR senses 2-methylisocitrate levels to regulate bacterial virulence in Pseudomonas aeruginosa.
Sci Adv, 8, 2022
|
|
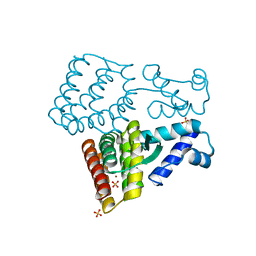
7XP0
 
 | |
7VR1
 
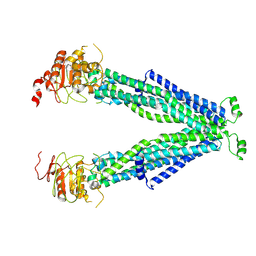 | |
7E5S
 
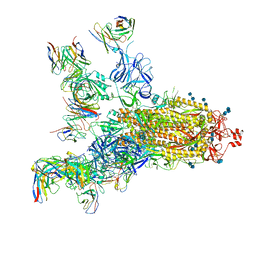 | | SARS-CoV-2 S trimer with four-antibody cocktail complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FC05 heavy chain, FC05 light chain, ... | | Authors: | Sun, Y, Wang, L, Wang, N, Feng, R, Wang, X. | | Deposit date: | 2021-02-20 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure-based development of three- and four-antibody cocktails against SARS-CoV-2 via multiple mechanisms.
Cell Res., 31, 2021
|
|
5Y1H
 
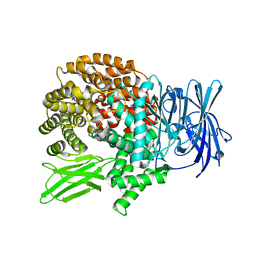 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,4-difluorobenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[[2,4-bis(fluoranyl)phenyl]methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-20 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,4-difluorobenzyl)ureido)-N-hydroxy-4-methylpentanamide
To Be Published
|
|
5Y1R
 
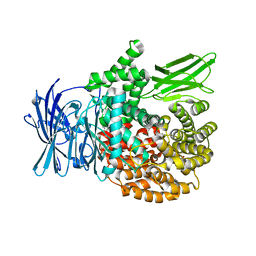 | |
5Y3I
 
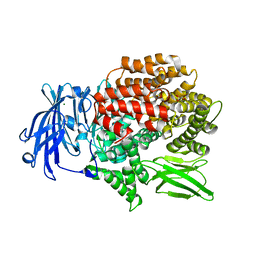 | |
5Y1S
 
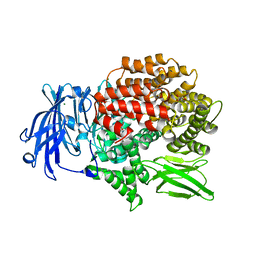 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3,4-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[(3,4-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, (2S)-2-[(3,4-dimethylphenyl)methylcarbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(3,4-dimethylbenzyl)ureido)-N-hydroxy-4-methylpentanamide
To Be Published
|
|
5Y1V
 
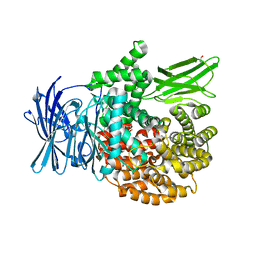 | | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide | | Descriptor: | (2R)-2-[(2,6-diethylphenyl)carbamoylamino]-4-methyl-N-oxidanyl-pentanamide, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Marapaka, A.K, Zhang, Y, Addlagatta, A. | | Deposit date: | 2017-07-21 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structure of Plasmodium falciparum aminopeptidase N in complex with (S)-2-(3-(2,6-diethylphenyl)ureido)- N-hydroxy-4-methylpentanamide
To Be Published
|
|
