8JH7
 
 | | FZD6 in inactive state | | Descriptor: | Frizzled-6,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-22 | | Release date: | 2024-01-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8J9N
 
 | | Gq bound FZD1 in ligand-free state | | Descriptor: | Frizzled-1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Lin, X, Xu, F. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8JHC
 
 | | FZD3 in inactive state | | Descriptor: | Frizzled-3,Soluble cytochrome b562, anti-BRIL Fab Heavy chain, anti-BRIL Fab Light chain, ... | | Authors: | Xu, F, Zhang, Z. | | Deposit date: | 2023-05-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8J9O
 
 | | Cryo-EM structure of inactive FZD1 | | Descriptor: | Frizzled-1,Soluble cytochrome b562, anti-BRIL Fab Heavy Chain, anti-BRIL Fab Light Chain, ... | | Authors: | Lin, X, Xu, F. | | Deposit date: | 2023-05-04 | | Release date: | 2024-01-17 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A framework for Frizzled-G protein coupling and implications to the PCP signaling pathways.
Cell Discov, 10, 2024
|
|
8JZE
 
 | | PSI-AcpPCI supercomplex from Symbiodinium | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z.H, Li, X.Y, Wang, W.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JW0
 
 | | PSI-AcpPCI supercomplex from Amphidinium carterae | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z.H, Li, X.Y, Wang, W.D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JZF
 
 | | PSI-AcpPCI supercomplex from Symbiodinium | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, X.Y, Li, Z.H, Wang, W.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8ZSP
 
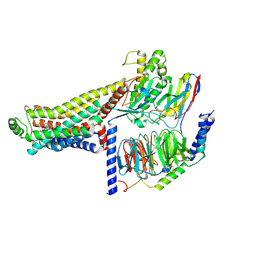 | | Cryo-EM structure of the LSD-bound hTAAR1-Gs complex | | Descriptor: | (8alpha)-N,N-diethyl-6-methyl-9,10-didehydroergoline-8-carboxamide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
8ZSV
 
 | | Cryo-EM structure of the RO5263397-bound mTAAR1-Gs complex | | Descriptor: | (4~{S})-4-(3-fluoranyl-2-methyl-phenyl)-1,3-oxazolidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
8ZSS
 
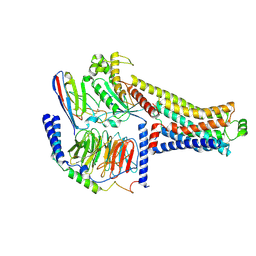 | | Cryo-EM structure of the RO5263397-bound hTAAR1-Gs complex | | Descriptor: | (4~{S})-4-(3-fluoranyl-2-methyl-phenyl)-1,3-oxazolidin-2-amine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
8ZSJ
 
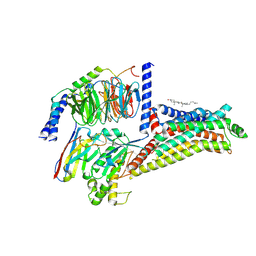 | | Cryo-EM structure of the apo hTAAR1-Gs complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Jiang, K.X, Zheng, Y, Xu, F. | | Deposit date: | 2024-06-05 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The versatile binding landscape of the TAAR1 pocket for LSD and other antipsychotic drug molecules.
Cell Rep, 43, 2024
|
|
7VF6
 
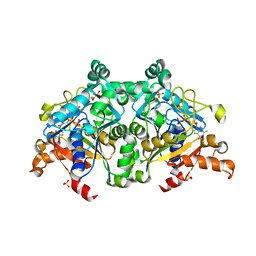 | | The crystal structure of PurZ0 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Tong, Y, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Alternative Z-genome biosynthesis pathway shows evolutionary progression from Archaea to phage.
Nat Microbiol, 8, 2023
|
|
7XCT
 
 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 2:1 complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XCR
 
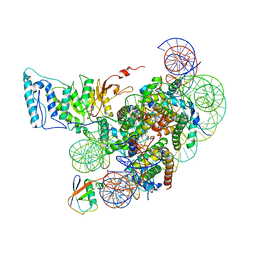 | | Cryo-EM structure of Dot1L and H2BK34ub-H3K79Nle nucleosome 1:1 complex | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-25 | | Release date: | 2022-04-20 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD0
 
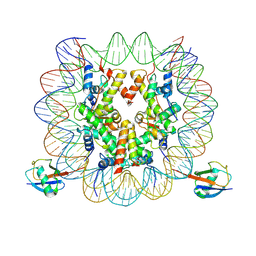 | | cryo-EM structure of H2BK34ub nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
7XD1
 
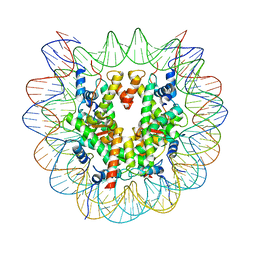 | | cryo-EM structure of unmodified nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, Liu, A.J, Lou, Z.Y, Liu, L. | | Deposit date: | 2022-03-26 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | H2B Lys34 Ubiquitination Induces Nucleosome Distortion to Stimulate Dot1L Activity.
Nat.Chem.Biol., 18, 2022
|
|
8GV3
 
 | | The cryo-EM structure of GSNOR with NYY001 | | Descriptor: | (4P)-4-{2-[4-(1H-imidazol-1-yl)phenyl]-5-[3-oxo-3-(2-oxo-1,3-thiazolidin-3-yl)propyl]-1H-pyrrol-1-yl}-3-methylbenzamide, Alcohol dehydrogenase class-3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Xia, Y, Zhang, Q, Yao, D, Zhao, S, Xie, L, Ji, Y, Cao, Y. | | Deposit date: | 2022-09-14 | | Release date: | 2023-09-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The cryo-EM structure of GSNOR with NYY001
To Be Published
|
|
5FT2
 
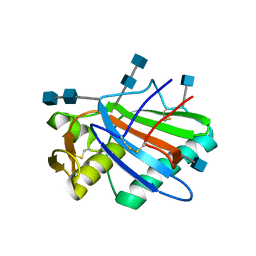 | | Sub-tomogram averaging of Lassa virus glycoprotein spike from virus- like particles at pH 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PRE-GLYCOPROTEIN POLYPROTEIN GP COMPLEX | | Authors: | Li, S, Zhaoyang, S, Pryce, R, Parsy, M.L, Fehling, S.K, Schlie, K, Siebert, C.A, Garten, W, Bowden, T.A, Strecker, T, Huiskonen, J.T. | | Deposit date: | 2016-01-09 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Acidic Ph-Induced Conformations and Lamp1 Binding of the Lassa Virus Glycoprotein Spike.
Plos Pathog., 12, 2016
|
|
