4GGN
 
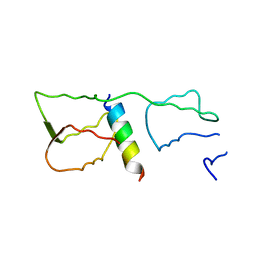 | | Malaria invasion machinery protein complex | | Descriptor: | Myosin A tail domain interacting protein MTIP, Myosin-A | | Authors: | Khamrui, S, Turley, S, Bergman, L.W, Hol, W.G.J. | | Deposit date: | 2012-08-06 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The structure of the D3 domain of Plasmodium falciparum myosin tail interacting protein MTIP in complex with a nanobody.
Mol.Biochem.Parasitol., 190, 2013
|
|
4GSP
 
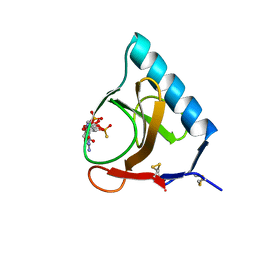 | | RIBONUCLEASE T1 COMPLEXED WITH 2',3'-CGPS + 3'-GMP, 7 DAYS | | Descriptor: | CALCIUM ION, GUANOSINE-2',3'-CYCLOPHOSPHOROTHIOATE, GUANOSINE-3'-MONOPHOSPHATE, ... | | Authors: | Zegers, I, Wyns, L. | | Deposit date: | 1997-12-02 | | Release date: | 1998-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analyzed by time-resolved crystallography.
Nat.Struct.Biol., 5, 1998
|
|
1LEM
 
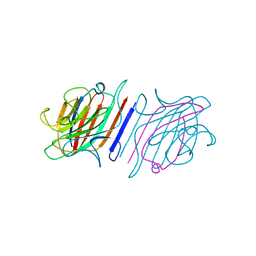 | |
6HHU
 
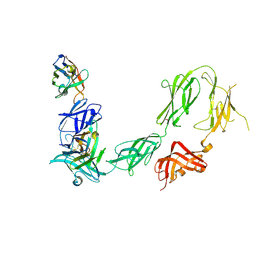 | |
5OCL
 
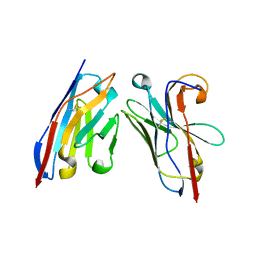 | | Nanobody-anti-VGLUT nanobody complex | | Descriptor: | anti-llama nanobody, anti-vglut nanobody | | Authors: | Dutzler, R, Schenck, S, Kunz, L. | | Deposit date: | 2017-07-03 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Generation and Characterization of Anti-VGLUT Nanobodies Acting as Inhibitors of Transport.
Biochemistry, 56, 2017
|
|
9G5K
 
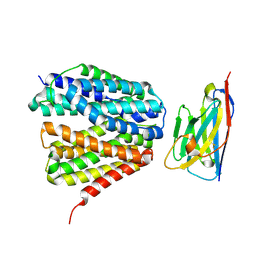 | |
1GSP
 
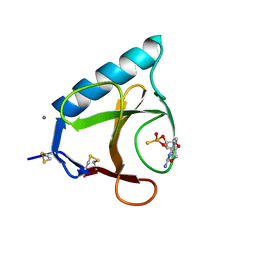 | | RIBONUCLEASE T1 COMPLEXED WITH 2',3'-CGPS, 1 DAY | | Descriptor: | CALCIUM ION, GUANOSINE-2',3'-CYCLOPHOSPHOROTHIOATE, RIBONUCLEASE T1 | | Authors: | Zegers, I, Wyns, L. | | Deposit date: | 1997-11-28 | | Release date: | 1998-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analyzed by time-resolved crystallography.
Nat.Struct.Biol., 5, 1998
|
|
9FYQ
 
 | | Cryo-EM structure of native SV2A in complex with TeNT-Hc, gangliosides and Pro-Macrobody 5 | | Descriptor: | (2~{R},4~{R},5~{S},6~{S})-2-[(2~{R},3~{R})-3-[(2~{R},3~{S},4~{S},6~{S})-6-[(2~{S},3~{S},4~{R},5~{S},6~{S})-3-[(2~{R},3~{R},4~{R},5~{S},6~{R})-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-2-(hydroxymethyl)-6-[(2~{R},3~{R},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-[2-(octadecanoylamino)-3-oxidanyl-octadec-4-enoxy]-4,5-bis(oxidanyl)oxan-3-yl]oxy-5-oxidanyl-oxan-4-yl]oxy-3-azanyl-6-carboxy-4-oxidanyl-oxan-2-yl]-2,3-bis(oxidanyl)propoxy]-5-azanyl-4-oxidanyl-6-[(1~{R},2~{S})-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, (2~{S},4~{S},5~{R},6~{S})-5-acetamido-2-[(2~{R},3~{R},4~{R},5~{S},6~{R})-2-[(2~{R},3~{R},4~{S},5~{R},6~{S})-2-[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(2~{S},4~{S},5~{S},6~{S})-5-acetamido-2-carboxy-4-oxidanyl-6-[(1~{S},2~{R})-1,2,3-tris(oxidanyl)propyl]oxan-2-yl]oxy-6-[(2~{R},3~{S},4~{R},5~{R},6~{R})-6-[2-(docosanoylamino)-3-oxidanyl-octadec-4-enoxy]-2-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-3-yl]oxy-2-(hydroxymethyl)-5-oxidanyl-oxan-3-yl]oxy-6-(hydroxymethyl)-5-oxidanyl-3-(2-oxidanylidenepropyl)oxan-4-yl]oxy-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-4-yl]oxy-4-oxidanyl-6-[(1~{R},2~{R})-1,2,3-tris(oxidanyl)propyl]oxane-2-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Schenck, S, Brunner, J.D. | | Deposit date: | 2024-07-03 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structures of native SV2A reveal the binding mode for tetanus neurotoxin and anti-epileptic racetams.
Nat Commun, 16, 2025
|
|
9FYR
 
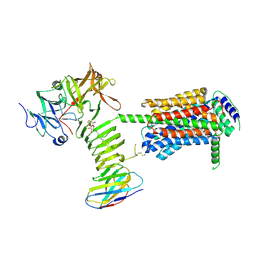 | |
9G97
 
 | | Lipid III flippase WzxE with NB10 nanobody in outward-facing conformation at 0.9688 A | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CHLORIDE ION, Lipid III flippase, ... | | Authors: | Le Bas, A, El Omari, K, Lee, M, Naismith, J.H. | | Deposit date: | 2024-07-24 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of WzxE the lipid III flippase for Enterobacterial Common Antigen polysaccharide.
Open Biology, 15, 2025
|
|
9G9N
 
 | |
9G95
 
 | | Lipid III flippase WzxE with NB10 nanobody in outward-facing conformation at 2.7552 A | | Descriptor: | CHLORIDE ION, Lipid III flippase, N-OCTANE, ... | | Authors: | Le Bas, A, El Omari, K, Lee, M, Naismith, J.H. | | Deposit date: | 2024-07-24 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of WzxE the lipid III flippase for Enterobacterial Common Antigen polysaccharide.
Open Biology, 15, 2025
|
|
9G9P
 
 | |
9G9M
 
 | |
9G9O
 
 | |
6QX4
 
 | |
1Q8F
 
 | |
5ICJ
 
 | | Crystal structure of the Mycobacterium tuberculosis transcriptional repressor EthR2 in complex with BDM41420 | | Descriptor: | 4,4,4-trifluoro-1-(3-phenyl-1-oxa-2,8-diazaspiro[4.5]dec-2-en-8-yl)butan-1-one, Probable transcriptional regulatory protein | | Authors: | Wohlkonig, A, Remaut, H, Tanina, A, Meyer, F, Willand, N, Baulard, A.R, Wintjens, R. | | Deposit date: | 2016-02-23 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversion of antibiotic resistance in Mycobacterium tuberculosis by spiroisoxazoline SMARt-420.
Science, 355, 2017
|
|
1LES
 
 | | LENTIL LECTIN COMPLEXED WITH SUCROSE | | Descriptor: | CALCIUM ION, LENTIL LECTIN, MANGANESE (II) ION, ... | | Authors: | Hamelryck, T, Loris, R. | | Deposit date: | 1995-08-23 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NMR, molecular modeling, and crystallographic studies of lentil lectin-sucrose interaction.
J.Biol.Chem., 270, 1995
|
|
1LEN
 
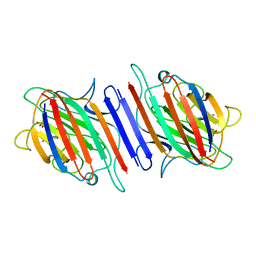 | | REFINEMENT OF TWO CRYSTAL FORMS OF LENTIL LECTIN AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Van Overberge, D, Loris, R, Wyns, L. | | Deposit date: | 1993-11-17 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of two crystal forms of lentil lectin at 1.8 A resolution.
Proteins, 20, 1994
|
|
7NHX
 
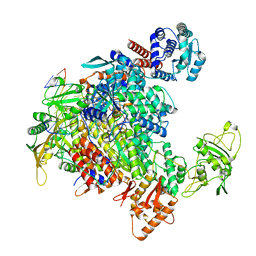 | | 1918 H1N1 Viral influenza polymerase heterotrimer - full transcriptase (Class1) | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, RNA (5'-R(P*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*CP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NKI
 
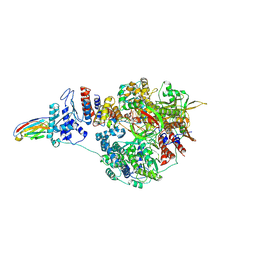 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8209 core | | Descriptor: | Nb8209, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.67 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NIS
 
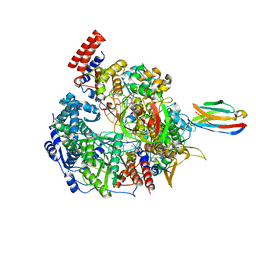 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8192 core | | Descriptor: | Nanobody8192 core, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-13 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.96 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NK1
 
 | | 1918 Influenza virus polymerase heterotirmer in complex with vRNA promoters and Nb8201 | | Descriptor: | Nanobody8201, Polymerase acidic protein, Polymerase basic protein 2,Immunoglobulin G-binding protein A, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-17 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
7NKR
 
 | | 1918 H1N1 Viral influenza polymerase heterotrimer with Nb8210 | | Descriptor: | Nb8210, Polymerase acidic protein, Polymerase basic protein 2,Polymerase basic protein 2, ... | | Authors: | Keown, J.R, Carrique, L, Fodor, E, Grimes, J.M. | | Deposit date: | 2021-02-18 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies.
Nat Commun, 13, 2022
|
|
