7E9P
 
 | |
7ENG
 
 | |
7E9N
 
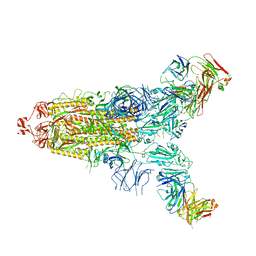 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with 35B5 Fab(1 down RBD, state1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 35B5 Fab, Light chain of 35B5 Fab, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-03-04 | | Release date: | 2022-04-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
7ENF
 
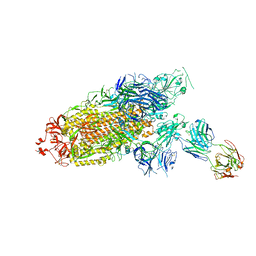 | | Cryo-EM structure of the SARS-CoV-2 S-6P in complex with Fab30 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab30, ... | | Authors: | Wang, X.F, Zhu, Y.Q. | | Deposit date: | 2021-04-16 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | A potent human monoclonal antibody with pan-neutralizing activities directly dislocates S trimer of SARS-CoV-2 through binding both up and down forms of RBD
Signal Transduct Target Ther, 7, 2022
|
|
8IHO
 
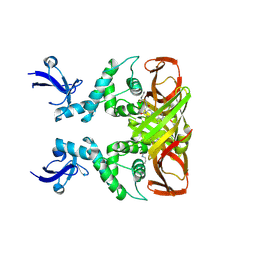 | | Crystal structures of SARS-CoV-2 papain-like protease in complex with covalent inhibitors | | Descriptor: | Papain-like protease nsp3, ZINC ION, covalent inhibitor | | Authors: | Wang, Q, Hu, H, Li, M, Xu, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure-Based Design of Potent Peptidomimetic Inhibitors Covalently Targeting SARS-CoV-2 Papain-like Protease.
Int J Mol Sci, 24, 2023
|
|
7ERY
 
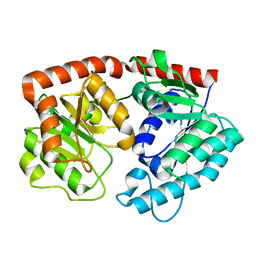 | | apo form of the glycosyltransferase | | Descriptor: | Glycosyltransferase | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES2
 
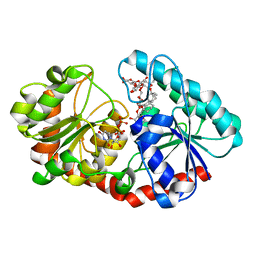 | | a mutant of glycosyktransferase in complex with UDP and Reb D | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, rebaudioside D | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ERX
 
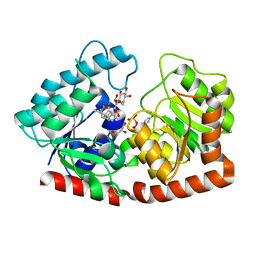 | | Glycosyltransferase in complex with UDP and STB | | Descriptor: | GLYCEROL, Glycosyltransferase, Steviolbioside, ... | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES1
 
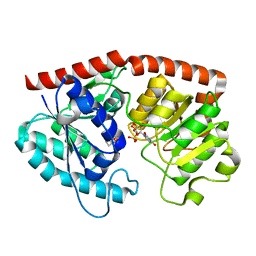 | | glycosyltransferase in complex with UDP and ST | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, steviol-19-o-glucoside | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
7ES0
 
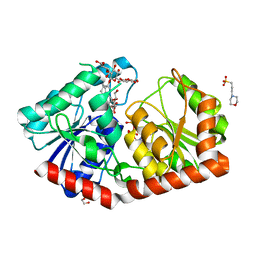 | | a rice glycosyltransferase in complex with UDP and REX | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, ... | | Authors: | Zhu, X. | | Deposit date: | 2021-05-08 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Catalytic flexibility of rice glycosyltransferase OsUGT91C1 for the production of palatable steviol glycosides.
Nat Commun, 12, 2021
|
|
5UJX
 
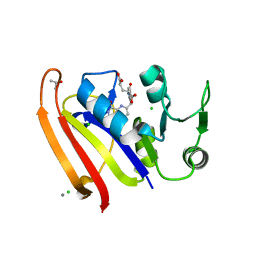 | | Crystal structure of DHFR in 20% Isopropanol | | Descriptor: | CALCIUM ION, CHLORIDE ION, Dihydrofolate reductase, ... | | Authors: | Cuneo, M.J, Agarwal, P.K. | | Deposit date: | 2017-01-19 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulating Enzyme Activity by Altering Protein Dynamics with Solvent.
Biochemistry, 57, 2018
|
|
2LKH
 
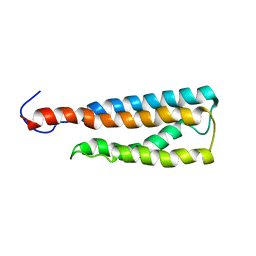 | | WSA minor conformation | | Descriptor: | Acetylcholine receptor | | Authors: | Xu, Y, Mowrey, D, Cui, T, Perez-Aguilar, J, Saven, J.G, Eckenhoff, R, Tang, P. | | Deposit date: | 2011-10-11 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of a designed water-soluble transmembrane domain of nicotinic acetylcholine receptor.
Biochim.Biophys.Acta, 1818, 2011
|
|
5HTL
 
 | | Structure of MshE with cdg | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), MSHA biogenesis protein MshE | | Authors: | Chin, K.H, Wang, Y.C. | | Deposit date: | 2016-01-27 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | Nucleotide binding by the widespread high-affinity cyclic di-GMP receptor MshEN domain.
Nat Commun, 7, 2016
|
|
4P1J
 
 | | Crystal structure of wild type Hypocrea jecorina Cel7a in a hexagonal crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exoglucanase 1, SAMARIUM (III) ION, ... | | Authors: | Bodenheimer, A.B, Cuneo, M.J, Swartz, P.D, Myles, D.A, Meilleur, F. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of wild type Hypocrea jecorina Cel7a in a hexagonal crystal form
To Be Published
|
|
4PPM
 
 | | Crystal structure of PigE: a transaminase involved in the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP) from Serratia sp. FS14 | | Descriptor: | Aminotransferase, MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Lou, X.D, Ran, T.T, Xu, D.Q, Wang, W.W. | | Deposit date: | 2014-02-27 | | Release date: | 2015-01-14 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the catalytic domain of PigE: a transaminase involved in the biosynthesis of 2-methyl-3-n-amyl-pyrrole (MAP) from Serratia sp. FS14
Biochem.Biophys.Res.Commun., 447, 2014
|
|
8YE9
 
 | |
8YE6
 
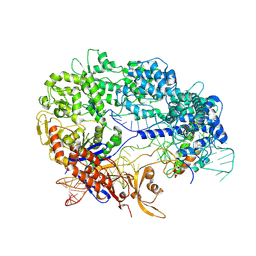 | |
7V3T
 
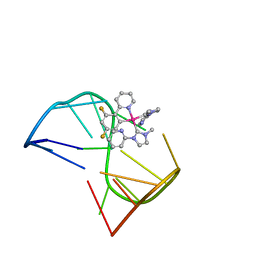 | | Solution structure of thrombin binding aptamer G-quadruplex bound a self-adaptive small molecule with rotated ligands | | Descriptor: | 11,13-bis(fluoranyl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-8-(1-methyl-3-pyridin-2-yl-imidazol-2-yl)-7$l^{4}-aza-8$l^{4}-platinatricyclo[7.4.0.0^{2,7}]trideca-1(9),2(7),3,5,10,12-hexaene, TBA G4 DNA (5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*GP*TP*TP*GP*G)-3') | | Authors: | Liu, W, Zhu, B.C, Mao, Z.W. | | Deposit date: | 2021-08-11 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a thrombin binding aptamer complex with a non-planar platinum(ii) compound.
Chem Sci, 13, 2022
|
|
8ZTO
 
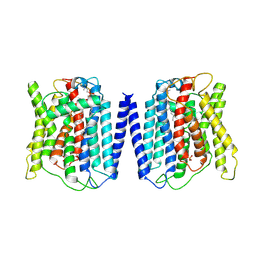 | | Cryo-EM structure of the human XPR1 | | Descriptor: | PHOSPHATE ION, Solute carrier family 53 member 1, [(2~{R})-1-hexadecanoyloxy-3-[oxidanyl-[2-(trimethyl-$l^{4}-azanyl)ethoxy]phosphoryl]oxy-propan-2-yl] octadec-9-enoate | | Authors: | She, J, Chen, L. | | Deposit date: | 2024-06-07 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structure and function of human XPR1 in phosphate export.
Nat Commun, 16, 2025
|
|
8XXT
 
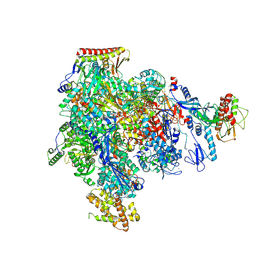 | | ASFV RNAP M1249L C-tail occupied complex2 (MCOC2) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XXP
 
 | | ASFV RNAP core complex | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-18 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XX4
 
 | | ASFV RNAP elongation complex | | Descriptor: | C122R, D339L, DNA (5'-D(P*CP*GP*CP*AP*AP*CP*GP*GP*CP*GP*A)-3'), ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8Y0E
 
 | | ASFV RNAP M1249L C-tail occupied complex4 (MCOC4) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-22 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XX5
 
 | | ASFV RNAP M1249L C-tail occupied complex1 (MCOC1) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-17 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
8XY6
 
 | | ASFV RNAP M1249L C-tail occupied complex3 (MCOC3) | | Descriptor: | C122R, C147L, D339L, ... | | Authors: | Zhu, G.L, Zhu, Y, Zhu, Z.X, Sun, F, Zheng, H.X. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of RNA polymerase complexes in African swine fever virus.
Nat Commun, 16, 2025
|
|
