7OIS
 
 | | mPI3Kd in complex with compound 7 | | Descriptor: | N-[5-[2-[(1S)-1-cyclopropylethyl]-7-[(3-methylsulfonylphenyl)sulfamoyl]-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION | | Authors: | Petersen, J. | | Deposit date: | 2021-05-12 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
4JIC
 
 | | Glycerol Trinitrate Reductase NerA from Agrobacterium radiobacter | | Descriptor: | DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, GTN Reductase, ... | | Authors: | Oberdorfer, G, Gruber, K. | | Deposit date: | 2013-03-05 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Structure of Glycerol Trinitrate Reductase NerA from Agrobacterium radiobacter Reveals the Molecular Reason for Nitro- and Ene-Reductase Activity in OYE Homologues.
Chembiochem, 14, 2013
|
|
4JIP
 
 | |
4JIQ
 
 | |
3IIT
 
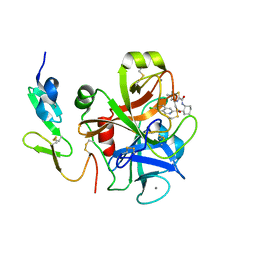 | | Factor XA in complex with a cis-1,2-diaminocyclohexane derivative | | Descriptor: | 7-chloro-N-[(1S,2R,4S)-4-(dimethylcarbamoyl)-2-{[(5-methyl-5,6-dihydro-4H-pyrrolo[3,4-d][1,3]thiazol-2-yl)carbonyl]amino}cyclohexyl]isoquinoline-3-carboxamide, Activated factor Xa heavy chain, CALCIUM ION, ... | | Authors: | Suzuki, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis, and SAR of cis-1,2-diaminocyclohexane derivatives as potent factor Xa inhibitors. Part II: exploration of 6-6 fused rings as alternative S1 moieties.
Bioorg.Med.Chem., 17, 2009
|
|
4XK0
 
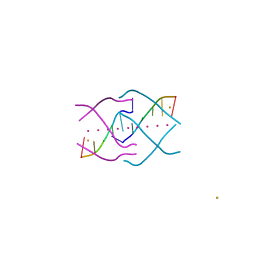 | | Crystal structure of a tetramolecular RNA G-quadruplex in potassium | | Descriptor: | BARIUM ION, POTASSIUM ION, RNA (5'-(*UP*GP*GP*GP*GP*U)-3') | | Authors: | Chen, M.C, Murat, P, Abecassis, K.A, Ferre-D'Amare, A.R, Balasubramanian, S. | | Deposit date: | 2015-01-09 | | Release date: | 2015-02-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Insights into the mechanism of a G-quadruplex-unwinding DEAH-box helicase.
Nucleic Acids Res., 43, 2015
|
|
4RZM
 
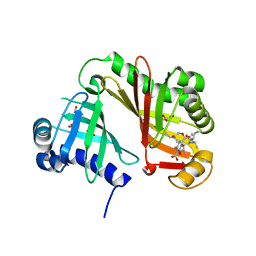 | | Crystal structure of the Lsd19-lasalocid A complex | | Descriptor: | CHLORIDE ION, Epoxide hydrolase LasB, FORMIC ACID, ... | | Authors: | Mathews, I.I, Hotta, K, Chen, X, Kim, C.-Y. | | Deposit date: | 2014-12-22 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Epoxide hydrolase-lasalocid a structure provides mechanistic insight into polyether natural product biosynthesis.
J.Am.Chem.Soc., 137, 2015
|
|
5NCZ
 
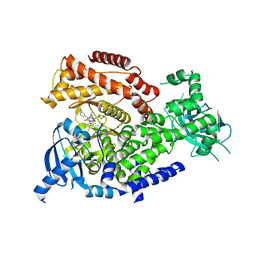 | | mPI3Kd IN COMPLEX WITH inh1 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-6-[[(1~{S})-1-[6-[3-[(dimethylamino)methyl]phenyl]-3-methyl-5-oxidanylidene-[1,3]thiazolo[3,2-a]pyridin-7-yl]ethyl]amino]pyrimidine-5-carbonitrile, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J. | | Deposit date: | 2017-03-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Design and Synthesis of Soluble and Cell-Permeable PI3K delta Inhibitors for Long-Acting Inhaled Administration.
J. Med. Chem., 60, 2017
|
|
1HMK
 
 | | RECOMBINANT GOAT ALPHA-LACTALBUMIN | | Descriptor: | CALCIUM ION, PROTEIN (ALPHA-LACTALBUMIN) | | Authors: | Horii, K, Matsushima, M, Tsumoto, K, Kumagai, I. | | Deposit date: | 1998-11-26 | | Release date: | 1999-11-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effect of the extra n-terminal methionine residue on the stability and folding of recombinant alpha-lactalbumin expressed in Escherichia coli.
J.Mol.Biol., 285, 1999
|
|
1WU1
 
 | |
2VC6
 
 | | Structure of MosA from S. meliloti with pyruvate bound | | Descriptor: | DIHYDRODIPICOLINATE SYNTHASE | | Authors: | Phenix, C.P, Nienaber, K.H, Tam, P.H, Delbaere, L.T.J, Palmer, D.R.J. | | Deposit date: | 2007-09-18 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural, functional and calorimetric investigation of MosA, a dihydrodipicolinate synthase from Sinorhizobium meliloti l5-30, does not support involvement in rhizopine biosynthesis.
Chembiochem, 9, 2008
|
|
7EF9
 
 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form II) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*AP*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
7EF8
 
 | | Crystal structure of mouse MUTYH in complex with DNA containing AP site analogue:8-oxoG (Form I) | | Descriptor: | Adenine DNA glycosylase, DNA (5'-D(*TP*AP*GP*TP*CP*CP*CP*(3DR)P*GP*TP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*AP*CP*(8OG)P*GP*GP*GP*AP*CP*T)-3'), ... | | Authors: | Nakamura, T, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2021-03-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of the mammalian adenine DNA glycosylase MUTYH: insights into the base excision repair pathway and cancer.
Nucleic Acids Res., 49, 2021
|
|
7EFA
 
 | |
7E5V
 
 | | Crystal structure of Phm7 in complex with inhibitor | | Descriptor: | Diels-Alderase, GLYCEROL, SULFATE ION, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E5U
 
 | | Crystal structure of Phm7 | | Descriptor: | CHLORIDE ION, Diels-Alderase, GLYCEROL, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E5T
 
 | | Crystal structure of Fsa2 | | Descriptor: | Diels-Alderase fsa2, ETHANOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16977525 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
4INS
 
 | | THE STRUCTURE OF 2ZN PIG INSULIN CRYSTALS AT 1.5 ANGSTROMS RESOLUTION | | Descriptor: | INSULIN (CHAIN A), INSULIN (CHAIN B), ZINC ION | | Authors: | Dodson, G.G, Dodson, E.J, Hodgkin, D.C, Isaacs, N.W, Vijayan, M. | | Deposit date: | 1989-07-10 | | Release date: | 1990-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of 2Zn pig insulin crystals at 1.5 A resolution.
Philos.Trans.R.Soc.London,Ser.B, 319, 1988
|
|
7A1A
 
 | |
7A19
 
 | |
1V3X
 
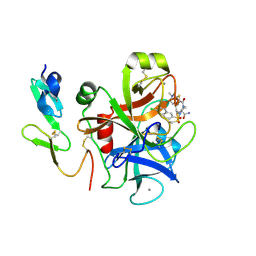 | | Factor Xa in complex with the inhibitor 1-[6-methyl-4,5,6,7-tetrahydrothiazolo(5,4-c)pyridin-2-yl] carbonyl-2-carbamoyl-4-(6-chloronaphth-2-ylsulphonyl)piperazine | | Descriptor: | (2R)-4-[(6-CHLORO-2-NAPHTHYL)SULFONYL]-1-[(5-METHYL-4,5,6,7-TETRAHYDRO[1,3]THIAZOLO[5,4-C]PYRIDIN-2-YL)CARBONYL]PIPERAZ INE-2-CARBOXAMIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Suzuki, M. | | Deposit date: | 2003-11-07 | | Release date: | 2004-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and conformational analysis of a non-amidine factor Xa inhibitor that incorporates 5-methyl-4,5,6,7-tetrahydrothiazolo[5,4-c]pyridine as S4 binding element
J.Med.Chem., 47, 2004
|
|
6A1G
 
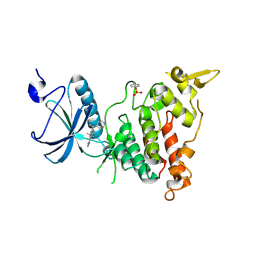 | | Crystal structure of human DYRK1A in complex with compound 32 | | Descriptor: | 5,5-dimethyl-8-[1-(piperidin-4-yl)ethenyl]-5,6-dihydrobenzo[h]quinazolin-4-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A | | Authors: | Baba, D, Hanzawa, H. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of DS42450411 as a potent orally active hepcidin production inhibitor: Design and optimization of novel 4-aminopyrimidine derivatives.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6A1F
 
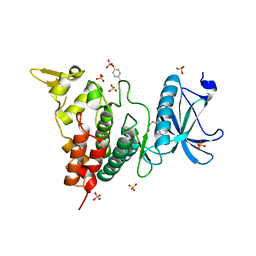 | | Crystal structure of human DYRK1A in complex with compound 14 | | Descriptor: | 8-methoxy-5,5-dimethyl-5,6-dihydrobenzo[h]quinazolin-4-amine, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, SULFATE ION, ... | | Authors: | Baba, D, Hanzawa, H. | | Deposit date: | 2018-06-07 | | Release date: | 2018-10-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of DS42450411 as a potent orally active hepcidin production inhibitor: Design and optimization of novel 4-aminopyrimidine derivatives.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5EMI
 
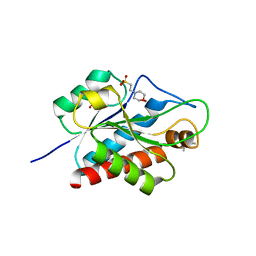 | | N-acetylmuramoyl-L-alanine amidase AmiC2 of Nostoc punctiforme | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cell wall hydrolase/autolysin, ... | | Authors: | Buettner, F.M, Stehle, T. | | Deposit date: | 2015-11-06 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Enabling cell-cell communication via nanopore formation: structure, function and localization of the unique cell wall amidase AmiC2 of Nostoc punctiforme.
Febs J., 283, 2016
|
|
4Z16
 
 | | Crystal Structure of the Jak3 Kinase Domain Covalently Bound to N-(3-(((5-chloro-2-((2-methoxy-4-(4-methylpiperazin-1-yl)phenyl)amino)pyrimidin-4-yl)amino)methyl)phenyl)acrylamide | | Descriptor: | N-(3-{[(5-chloro-2-{[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]methyl}phenyl)prop-2-enamide, Tyrosine-protein kinase JAK3 | | Authors: | McNally, R, Tan, L, Gray, N.S, Eck, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-02-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Development of Selective Covalent Janus Kinase 3 Inhibitors.
J.Med.Chem., 58, 2015
|
|
