8F98
 
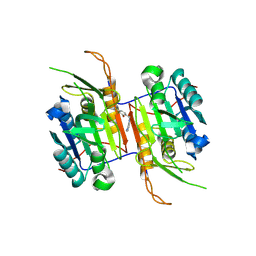 | | Compound 8 bound to procaspase-6 | | Descriptor: | 5-fluoro-2-({[(3M)-3-(1H-imidazol-4-yl)pyridin-2-yl]amino}methyl)phenol, Procaspase-6 | | Authors: | Fan, P, Zhao, Y, Renslo, A.R, Arkin, M.R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Study of Heteroarene Stacking Using a Congeneric Set of Molecular Glues for Procaspase-6.
J.Med.Chem., 66, 2023
|
|
8F96
 
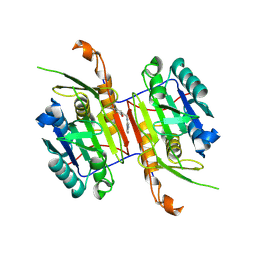 | | Compound 3 bound to procaspase-6 | | Descriptor: | 5-fluoro-2-({[(3M)-3-(pyrimidin-4-yl)pyridin-2-yl]amino}methyl)phenol, Procaspase-6 | | Authors: | Fan, P, Zhao, Y, Renslo, A.R, Arkin, M.R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Systematic Study of Heteroarene Stacking Using a Congeneric Set of Molecular Glues for Procaspase-6.
J.Med.Chem., 66, 2023
|
|
8F78
 
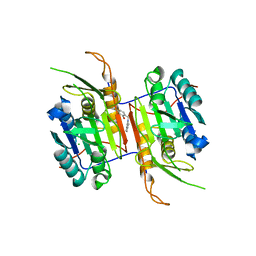 | | Compound 1 bound to procaspase-6 | | Descriptor: | 5-fluoro-2-({[3-(pyrimidin-2-yl)pyridin-2-yl]amino}methyl)phenol, CHLORIDE ION, Procaspase-6 | | Authors: | Fan, P, Zhao, Y, Renslo, A.R, Arkin, M.R. | | Deposit date: | 2022-11-18 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Systematic Study of Heteroarene Stacking Using a Congeneric Set of Molecular Glues for Procaspase-6.
J.Med.Chem., 66, 2023
|
|
8FBV
 
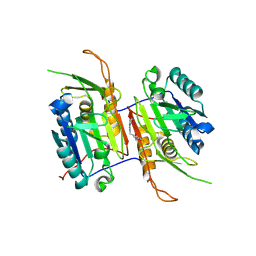 | | Compound 7 bound to procaspase-6 | | Descriptor: | 5-fluoro-2-({[(3M)-3-(1H-imidazol-2-yl)pyridin-2-yl]amino}methyl)phenol, Procaspase-6 | | Authors: | Fan, P, Zhao, Y, Renslo, A.R, Arkin, M.R. | | Deposit date: | 2022-11-30 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Systematic Study of Heteroarene Stacking Using a Congeneric Set of Molecular Glues for Procaspase-6.
J.Med.Chem., 66, 2023
|
|
7BR2
 
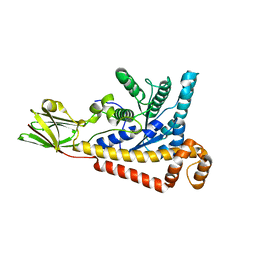 | |
1LS4
 
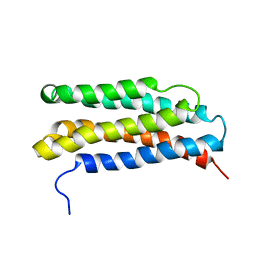 | |
6A98
 
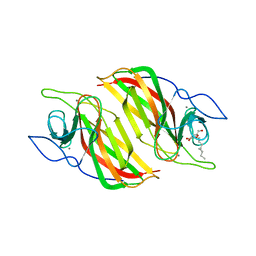 | | Crystal structure of a cyclase from Fischerella sp. TAU | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, aromatic prenyltransferase, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6A8X
 
 | | Crystal structure of a apo form cyclase from Fischerella sp. | | Descriptor: | CALCIUM ION, aromatic prenyltransferase | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-11 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6A92
 
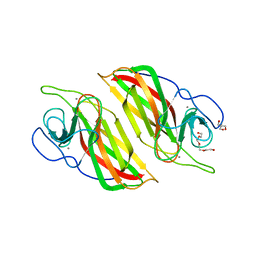 | | Crystal structure of a cyclase Filc1 from Fischerella sp. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-11 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6A9F
 
 | | Crystal structure of a cyclase from Fischerella sp. TAU in complex with 4-(1H-Indol-3-yl)butan-2-one | | Descriptor: | 4-(1~{H}-indol-3-yl)butan-2-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-13 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6A99
 
 | | Crystal structure of a Stig cyclases Fisc from Fischerella sp. TAU in complex with (3Z)-3-(1-methyl-2-pyrrolidinylidene)-3H-indole | | Descriptor: | (3~{Z})-3-(1-methylpyrrolidin-2-ylidene)indole, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Hu, X.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-07-12 | | Release date: | 2018-12-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6ADU
 
 | | Crystal structure of an enzyme in complex with ligand C | | Descriptor: | (3~{Z})-3-(1-methylpyrrolidin-2-ylidene)indole, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Tan, X.K, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-08-02 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The Crystal Structure of a Class of Cyclases that Catalyze the Cope Rearrangement.
Angew.Chem.Int.Ed.Engl., 57, 2018
|
|
6LON
 
 | | Crystal structure of HPSG | | Descriptor: | (2~{S})-2,3-bis(oxidanyl)propane-1-sulfonic acid, GLYCEROL, PFL2/glycerol dehydratase family glycyl radical enzyme, ... | | Authors: | Liu, J, Zhang, Y, Yuchi, Z. | | Deposit date: | 2020-01-06 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two radical-dependent mechanisms for anaerobic degradation of the globally abundant organosulfur compound dihydroxypropanesulfonate.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6KV9
 
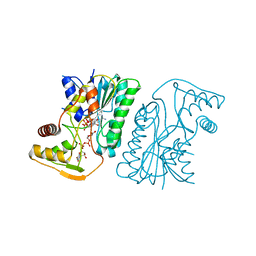 | | MoeE5 in complex with UDP-glucuronic acid and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
5HCV
 
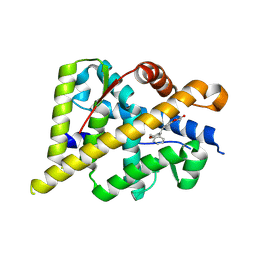 | |
4FMA
 
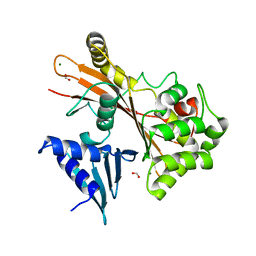 | | EspG structure | | Descriptor: | ACETIC ACID, EspG protein, FORMIC ACID, ... | | Authors: | Shao, F, Zhu, Y, Hu, L. | | Deposit date: | 2012-06-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structurally Distinct Bacterial TBC-like GAPs Link Arf GTPase to Rab1 Inactivation to Counteract Host Defenses.
Cell(Cambridge,Mass.), 150, 2012
|
|
3PDU
 
 | |
3PEF
 
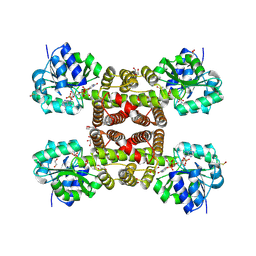 | |
5IO1
 
 | |
5INW
 
 | | Structure of reaction loop cleaved lamprey angiotensinogen | | Descriptor: | C-terminal peptide of Putative angiotensinogen, Putative angiotensinogen, SULFATE ION | | Authors: | Wei, H, Zhou, A. | | Deposit date: | 2016-03-08 | | Release date: | 2016-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Heparin Binds Lamprey Angiotensinogen and Promotes Thrombin Inhibition through a Template Mechanism
J.Biol.Chem., 291, 2016
|
|
6D06
 
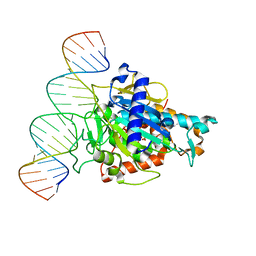 | | Human ADAR2d E488Y mutant complexed with dsRNA containing an abasic site opposite the edited base | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*CP*AP*GP*AP*GP*CP*CP*CP*CP*CP*NP*AP*GP*CP*AP*UP*CP*GP*CP*GP*AP*GP*C)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A Bump-Hole Approach for Directed RNA Editing.
Cell Chem Biol, 26, 2019
|
|
2H35
 
 | | Solution structure of Human normal adult hemoglobin | | Descriptor: | Hemoglobin alpha subunit, Hemoglobin beta subunit, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Fan, J.S, Yang, D. | | Deposit date: | 2006-05-22 | | Release date: | 2006-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A new strategy for structure determination of large proteins in solution without deuteration
Nat.Methods, 3, 2006
|
|
2L8L
 
 | |
5XFZ
 
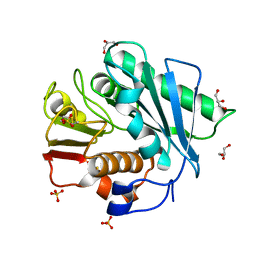 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant from Ideonella sakaiensis 201-F6 | | Descriptor: | GLYCEROL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
5XH2
 
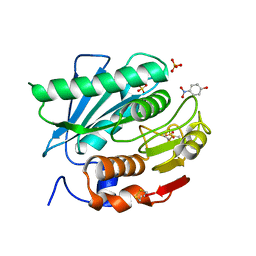 | | Crystal structure of a novel PET hydrolase R103G/S131A mutant in complex with pNP from Ideonella sakaiensis 201-F6 | | Descriptor: | P-NITROPHENOL, Poly(ethylene terephthalate) hydrolase, SULFATE ION | | Authors: | Han, X, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insight into catalytic mechanism of PET hydrolase
Nat Commun, 8, 2017
|
|
