7YMI
 
 | | PSII-Pcb Dimer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of a unique PSII-Pcb tetrameric megacomplex in a chlorophyll d -containing cyanobacterium.
Sci Adv, 10, 2024
|
|
7YMM
 
 | | PSII-Pcb Tetramer of Acaryochloris Marina | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (6'R,11cis,11'cis,13cis,15cis)-4',5'-didehydro-5',6'-dihydro-beta,beta-carotene, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Shen, L.L, Gao, Y.Z, Wang, W.D, Zhang, X, Shen, J.R, Wang, P.Y, Han, G.Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-08-16 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of a unique PSII-Pcb tetrameric megacomplex in a chlorophyll d -containing cyanobacterium.
Sci Adv, 10, 2024
|
|
8XUS
 
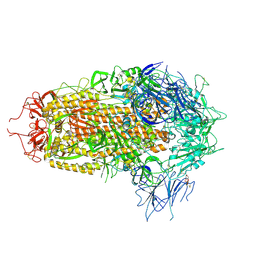 | | JN.1 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUT
 
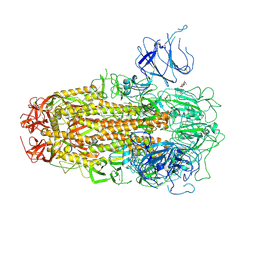 | | XBB.1.5 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P, Mao, X. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUR
 
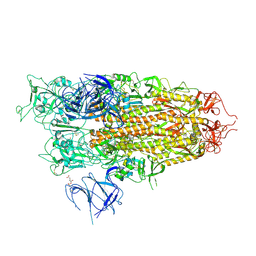 | | BA.2.86 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUU
 
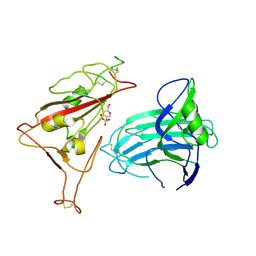 | | BA.2.86-T356K Spike Trimer in complex with heparan sulfate (Local refinement) | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8GQU
 
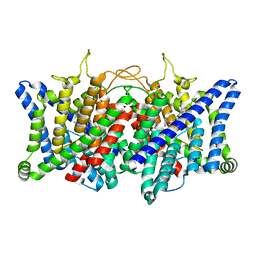 | | AK-42 inhibitor binding human ClC-2 TMD | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42.
Nat Commun, 14, 2023
|
|
7JVN
 
 | |
7JVM
 
 | |
8X4Z
 
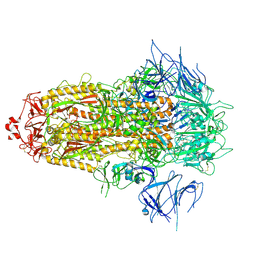 | | BA.2.86 Spike Trimer with ins483V mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X56
 
 | | BA.2.86 Spike Trimer with T356K mutation (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X55
 
 | | BA.2.86 Spike Trimer with T356K mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5R
 
 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X4H
 
 | | SARS-CoV-2 JN.1 Spike | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-15 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X5Q
 
 | | SARS-CoV-2 BA.2.75 Spike with K356T mutation (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-17 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8X50
 
 | | BA.2.86 Spike Trimer with ins483V mutation (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2023-11-16 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
5AZF
 
 | | Crystal structure of LGG-1 complexed with a WEEL peptide | | Descriptor: | CADMIUM ION, Protein lgg-1, SULFATE ION, ... | | Authors: | Watanabe, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
7WRJ
 
 | |
7WRY
 
 | |
5AZG
 
 | | Crystal structure of LGG-1 complexed with a UNC-51 peptide | | Descriptor: | CADMIUM ION, Protein lgg-1, Serine/threonine-protein kinase unc-51 | | Authors: | Watanabe, Y, Fujioka, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
5AZH
 
 | | Crystal structure of LGG-2 fused with an EEEWEEL peptide | | Descriptor: | EEEWEEL peptide,Protein lgg-2, MAGNESIUM ION | | Authors: | Watanabe, Y, Fujioka, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
7Y0W
 
 | | Local structure of BD55-5514 and BD55-5840 Fab and Omicron BA.1 RBD complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD55-5514H, BD55-5514L, ... | | Authors: | Zhang, Z, Xiao, J. | | Deposit date: | 2022-06-06 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Rational identification of potent and broad sarbecovirus-neutralizing antibody cocktails from SARS convalescents.
Cell Rep, 41, 2022
|
|
6LA3
 
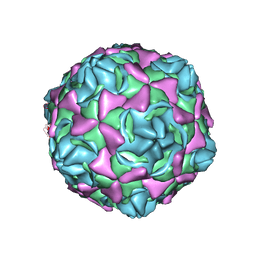 | | Cryo-EM structure of full echovirus 11 particle at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, S, Gao, F.G. | | Deposit date: | 2019-11-11 | | Release date: | 2020-10-07 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Molecular and structural basis of Echovirus 11 infection by using the dual-receptor system of CD55 and FcRn.
Chin.Sci.Bull., 2020
|
|
6LAO
 
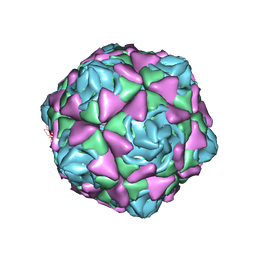 | |
6M7P
 
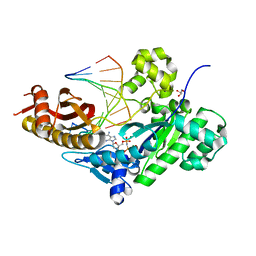 | | Human DNA polymerase eta extension complex with cdA at the -2 position | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*TP*GP*TP*GP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*TP*CP*(02I)P*CP*AP*CP*AP*CP*T)-3'), ... | | Authors: | Gregory, M.T, Yang, W. | | Deposit date: | 2018-08-20 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Bypassing a 8,5'-cyclo-2'-deoxyadenosine lesion by human DNA polymerase eta at atomic resolution.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
