4OCN
 
 | |
4OCL
 
 | |
4OCM
 
 | |
6QXE
 
 | | Influenza A virus (A/NT/60/1968) polymerase dimer of hetermotrimer in complex with 3'5' cRNA promoter and Nb8205 | | Descriptor: | Nb8205, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2019-03-07 | | Release date: | 2019-09-04 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structures of influenza A virus RNA polymerase offer insight into viral genome replication.
Nature, 573, 2019
|
|
5N7O
 
 | | EthR2 in complex with SMARt-420 compound | | Descriptor: | 4,4,4-trifluoro-1-(3-phenyl-1-oxa-2,8-diazaspiro[4.5]dec-2-en-8-yl)butan-1-one, Probable transcriptional regulatory protein | | Authors: | Wohlkonig, A, Wintjens, R. | | Deposit date: | 2017-02-21 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the interaction between spiroisoxazoline SMARt-420 and the Mycobacterium tuberculosis repressor EthR2.
Biochem. Biophys. Res. Commun., 487, 2017
|
|
8S5W
 
 | | Cryo-EM structure of fedratinib-bound human SLC19A3 in inward-open state | | Descriptor: | N-tert-butyl-3-{[5-methyl-2-({4-[2-(pyrrolidin-1-yl)ethoxy]phenyl}amino)pyrimidin-4-yl]amino}benzenesulfonamide, Nb3.7, Thiamine transporter 2 | | Authors: | Gabriel, F, Loew, C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of thiamine transport and drug recognition by SLC19A3.
Nat Commun, 15, 2024
|
|
8S5U
 
 | | Cryo-EM structure of thiamine-bound human SLC19A3 in outward-open state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, Nb3.4, ... | | Authors: | Gabriel, F, Loew, C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis of thiamine transport and drug recognition by SLC19A3.
Nat Commun, 15, 2024
|
|
8S5Z
 
 | |
8S61
 
 | |
8S4U
 
 | |
8S62
 
 | |
8RRS
 
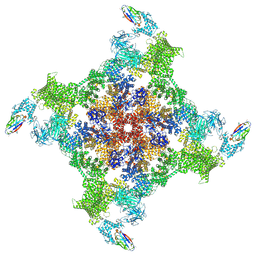 | | Structure of mouse RyR2 solubilised in detergent in open state in complex with Ca2+, ATP, caffeine and Nb9657. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, CALCIUM ION, ... | | Authors: | Li, C, Efremov, R.G. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM.
J.Biol.Chem., 300, 2024
|
|
8RRW
 
 | |
8RRV
 
 | |
8RRT
 
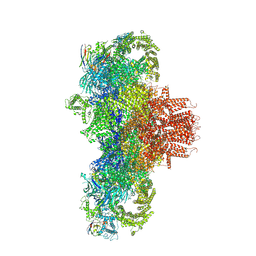 | |
8RRX
 
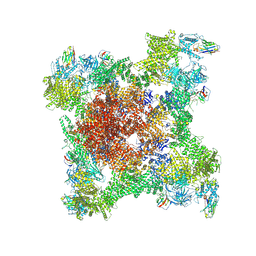 | | Structure of RyR1 reconstituted into lipid nanodisc in primed state in complex with Ca2+, ATP, caffeine and Nb9657 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Li, C, Efremov, R.G. | | Deposit date: | 2024-01-23 | | Release date: | 2024-10-09 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rapid small-scale nanobody-assisted purification of ryanodine receptors for cryo-EM.
J.Biol.Chem., 300, 2024
|
|
8RS0
 
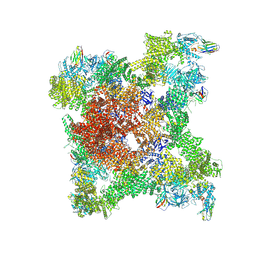 | |
8RRU
 
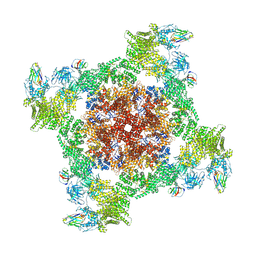 | |
4EIG
 
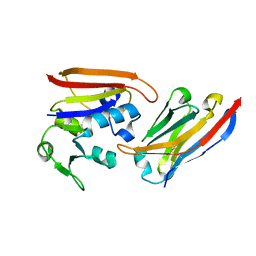 | | CA1698 camel antibody fragment in complex with DHFR | | Descriptor: | CA1698 camel antibody fragment, Dihydrofolate reductase | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-05 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
4EIZ
 
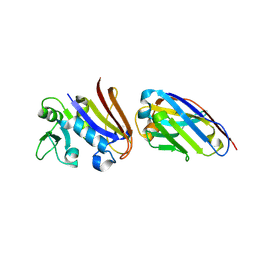 | | Structure of Nb113 bound to apoDHFR | | Descriptor: | Dihydrofolate reductase, Nb113 Camel antibody fragment | | Authors: | Oyen, D, Srinivasan, V. | | Deposit date: | 2012-04-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
9ENA
 
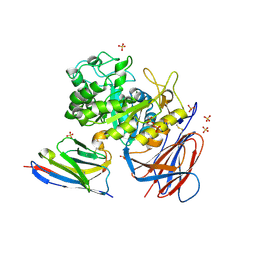 | | Lysosomal glucocerebrosidase in complex with a stabilizing nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chains: B, ... | | Authors: | Dal Maso, T, Versees, W. | | Deposit date: | 2024-03-12 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Developing nanobodies as allosteric molecular chaperones of glucocerebrosidase function.
Nat Commun, 16, 2025
|
|
6FPV
 
 | | A llama-derived JBP1-targeting nanobody | | Descriptor: | GLYCEROL, Nanobody | | Authors: | van Beusekom, B, Adamopoulos, A, Heidebrecht, T, Joosten, R.P, Perrakis, A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization and structure determination of a llama-derived nanobody targeting the J-base binding protein 1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
4EJ1
 
 | |
6GSP
 
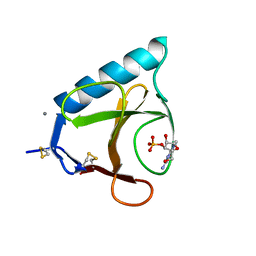 | | RIBONUCLEASE T1/3'-GMP, 15 WEEKS | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Zegers, I, Wyns, L. | | Deposit date: | 1997-12-09 | | Release date: | 1998-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hydrolysis of a slow cyclic thiophosphate substrate of RNase T1 analyzed by time-resolved crystallography.
Nat.Struct.Biol., 5, 1998
|
|
4FHB
 
 | | Enhancing DHFR catalysis by binding of an allosteric regulator nanobody (Nb179) | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Oyen, D. | | Deposit date: | 2012-06-06 | | Release date: | 2013-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic analysis of allosteric and non-allosteric effects arising from nanobody binding to two epitopes of the dihyrofolate reductase of Escherichia coli.
Biochim.Biophys.Acta, 1834, 2013
|
|
