3XIN
 
 | |
2XIN
 
 | |
4XIM
 
 | |
7AWP
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with rubidium and barium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.91 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWM
 
 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with L-ASP, three sodium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, BARIUM ION, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWL
 
 | | Structure of the thermostabilized EAAT1 cryst-II mutant in complex with barium and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWQ
 
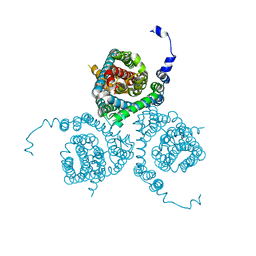 | | Structure of the thermostabilized EAAT1 cryst-E386Q mutant in complex with L-ASP, sodium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, BARIUM ION, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWN
 
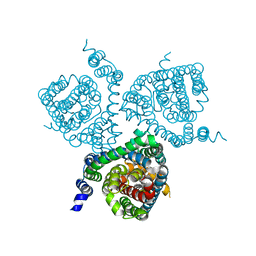 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with rubidium and barium and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7NPW
 
 | |
3SR2
 
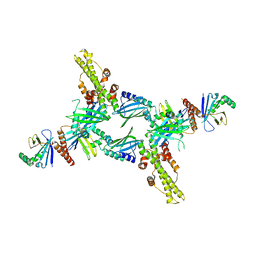 | | Crystal Structure of Human XLF-XRCC4 Complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Hammel, M, Classen, S, Tainer, J.A. | | Deposit date: | 2011-07-06 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.9708 Å) | | Cite: | XRCC4 Protein Interactions with XRCC4-like Factor (XLF) Create an Extended Grooved Scaffold for DNA Ligation and Double Strand Break Repair.
J.Biol.Chem., 286, 2011
|
|
6SK0
 
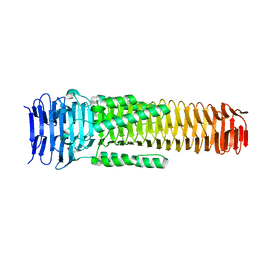 | | The VgrG spike from the Type 6 secretion system | | Descriptor: | Putative type VI secretion protein | | Authors: | Rapisarda, C, Fronzes, R. | | Deposit date: | 2019-08-14 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for loading and inhibition of a bacterial T6SS phospholipase effector by the VgrG spike.
Embo J., 39, 2020
|
|
6SKI
 
 | |
6SJL
 
 | |
7PDQ
 
 | | Crystal structure of a mutated form of RXRalpha ligand binding domain in complex with LG100268 and a coactivator fragment | | Descriptor: | 6-[1-(3,5,5,8,8-PENTAMETHYL-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)CYCLOPROPYL]PYRIDINE-3-CARBOXYLIC ACID, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | le Maire, A, Bourguet, W, Guee, L. | | Deposit date: | 2021-08-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
7PDT
 
 | | Crystal structure of a mutated form of RXRalpha ligand binding domain in complex with BMS649 and a coactivator fragment | | Descriptor: | 4-[2-(5,5,8,8-TETRAMETHYL-5,6,7,8-TETRAHYDRO-NAPHTHALEN-2-YL)-[1,3]DIOXOLAN-2-YL]-BENZOIC ACID, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | le Maire, A, Bourguet, W, Guee, L. | | Deposit date: | 2021-08-07 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
8P2V
 
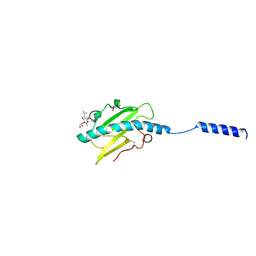 | | Neisseria meningitidis Type IV pilus SB-GATDH variant | | Descriptor: | (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methyl-4,6-bis(oxidanyl)oxan-3-yl]-2,3-bis(oxidanyl)propanamide, Neisseria meningitidis PilE variant SB-GATDH, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-05-16 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
8P36
 
 | | Neisseria meningitidis Type IV pilus SB-DATDH variant | | Descriptor: | 2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, Neisseria meningitidis PilE, SB-DATDH variant, ... | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-05-17 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
8PIJ
 
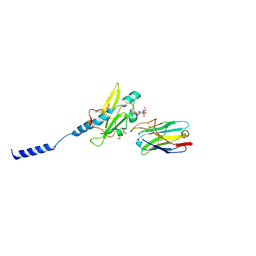 | | Neisseria meningitidis Type IV pilus SB-GATDH variant bound to the C24 nanobody | | Descriptor: | (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methyl-4,6-bis(oxidanyl)oxan-3-yl]-2,3-bis(oxidanyl)propanamide, C24 nanobody, Pilin, ... | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-06-21 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
8PJP
 
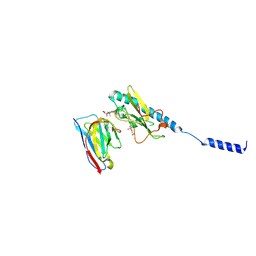 | | Neisseria meningitidis PilE, SB-GATDH variant, bound to the F10 nanobody | | Descriptor: | (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methyl-4,6-bis(oxidanyl)oxan-3-yl]-2,3-bis(oxidanyl)propanamide, Nanobody F10, Pilin, ... | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-06-23 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
8PIZ
 
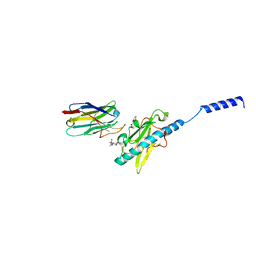 | | Neisseria meningitidis Type IV pilus SB-DATDH variant bound to the C24 nanobody | | Descriptor: | 2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, C24 nanobody, Pilin, ... | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-06-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
7QAA
 
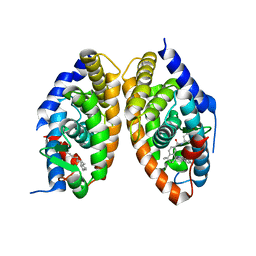 | | Crystal structure of RARalpha/RXRalpha ligand binding domain heterodimer in complex with BMS614 and oleic acid | | Descriptor: | 4-[(4,4-DIMETHYL-1,2,3,4-TETRAHYDRO-[1,2']BINAPTHALENYL-7-CARBONYL)-AMINO]-BENZOIC ACID, Isoform Alpha-1-deltaBC of Retinoic acid receptor alpha, OLEIC ACID, ... | | Authors: | le Maire, A, Vivat, V, Guee, L, Blanc, P, Malosse, C, Chamot-Rooke, J, Germain, P, Bourguet, w. | | Deposit date: | 2021-11-16 | | Release date: | 2022-10-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Design and in vitro characterization of RXR variants as tools to investigate the biological role of endogenous rexinoids.
J.Mol.Endocrinol., 69, 2022
|
|
8P3B
 
 | | Neisseria meningitidis Type IV pilus SA-GATDH variant | | Descriptor: | (2~{R})-~{N}-[(2~{R},3~{S},4~{S},5~{R},6~{R})-5-acetamido-2-methyl-4,6-bis(oxidanyl)oxan-3-yl]-2,3-bis(oxidanyl)propanamide, Fimbrial protein, SN-GLYCEROL-3-PHOSPHATE | | Authors: | Fernandez-Martinez, D, Dumenil, G. | | Deposit date: | 2023-05-17 | | Release date: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Cryo-EM structures of type IV pili complexed with nanobodies reveal immune escape mechanisms.
Nat Commun, 15, 2024
|
|
6GJ3
 
 | |
6GJ1
 
 | |
6GIY
 
 | |
