1EHG
 
 | |
1EHF
 
 | |
2LM4
 
 | | Solution NMR Structure of mitochondrial succinate dehydrogenase assembly factor 2 from Saccharomyces cerevisiae, Northeast Structural Genomics Consortium Target YT682A | | Descriptor: | Succinate dehydrogenase assembly factor 2, mitochondrial | | Authors: | Eletsky, A, Winge, D.R, Lee, H, Lee, D, Kohan, E, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-11-22 | | Release date: | 2012-01-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of yeast succinate dehydrogenase flavinylation factor sdh5 reveals a putative sdh1 binding site.
Biochemistry, 51, 2012
|
|
1EHE
 
 | |
2LRQ
 
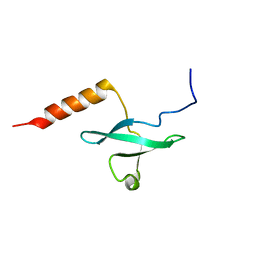 | | Chemical Shift Assignment and Solution Structure of Fr822A from Drosophila melanogaster. Northeast Structural Genomics Consortium Target Fr822A | | Descriptor: | NuA4 complex subunit EAF3 homolog | | Authors: | Lee, H, Lee, D, Kohan, E, Janjua, H, Xiao, R, Acton, T, Everett, J.K, Montelione, G, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG), Chaperone-Enabled Studies of Epigenetic Regulation Enzymes (CEBS) | | Deposit date: | 2012-04-11 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Fr822A from Drosophila melanogaster.
To be Published
|
|
2LEQ
 
 | | Chemical Shift Assignment and Solution Structure of ChR145 from Cytophaga Hutchinsonii, Northeast Structural Genomics Consortium Target ChR145 | | Descriptor: | Uncharacterized protein | | Authors: | Lee, H, Lee, D, Ciccosanti, C, Mao, L.R, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of ChR145.
To be Published
|
|
5IUZ
 
 | | STRUCTURE OF P450 2B4 F202W MUTANT (CYMAL-5) | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jang, H.-H, Halpert, J.R, Shah, M.B. | | Deposit date: | 2016-03-18 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Effect of detergent binding on cytochrome P450 2B4 structure as analyzed by X-ray crystallography and deuterium-exchange mass spectrometry.
Biophys.Chem., 216, 2016
|
|
4EJ0
 
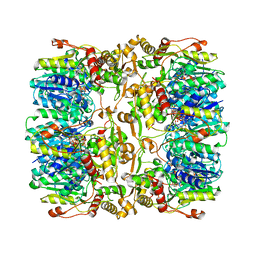 | |
1DMN
 
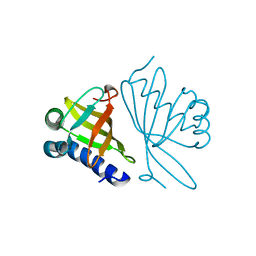 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F/Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMM
 
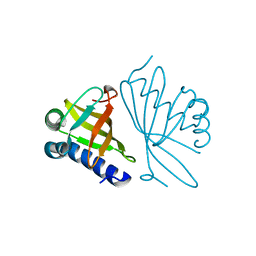 | | CRYSTAL STRUCTURES OF MUTANT ENZYMES Y57F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
1DMQ
 
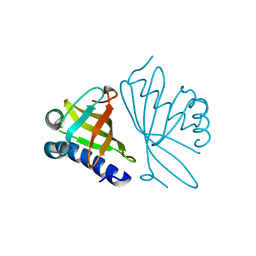 | | CRYSTAL STRUCTURE OF MUTANT ENZYME Y32F OF KETOSTEROID ISOMERASE FROM PSEUDOMONAS PUTIDA BIOTYPE B | | Descriptor: | STEROID DELTA-ISOMERASE | | Authors: | Kim, D.H, Jang, D.S, Nam, G.H, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-12-14 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Contribution of the hydrogen-bond network involving a tyrosine triad in the active site to the structure and function of a highly proficient ketosteroid isomerase from Pseudomonas putida biotype B.
Biochemistry, 39, 2000
|
|
5V2A
 
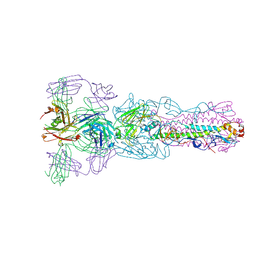 | | Crystal structure of Fab H7.167 in complex with influenza virus hemagglutinin from A/Shanghai/02/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H7.167 antibody, Hemagglutinin, ... | | Authors: | Zhang, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2017-03-03 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.656 Å) | | Cite: | H7N9 influenza virus neutralizing antibodies that possess few somatic mutations.
J. Clin. Invest., 126, 2016
|
|
5WOB
 
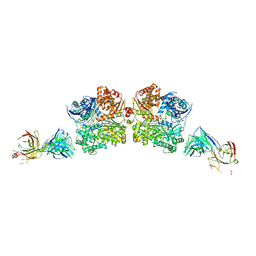 | | Crystal Structure Analysis of Fab1-Bound Human Insulin Degrading Enzyme (IDE) in Complex with Insulin | | Descriptor: | IDE-bound Fab heavy chain, IDE-bound Fab light chain, Insulin, ... | | Authors: | McCord, L.A, Liang, W.G, Farcasanu, M, Wang, A.G, Koide, S, Tang, W.J. | | Deposit date: | 2017-08-01 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Ensemble cryoEM elucidates the mechanism of insulin capture and degradation by human insulin degrading enzyme.
Elife, 7, 2018
|
|
2KYB
 
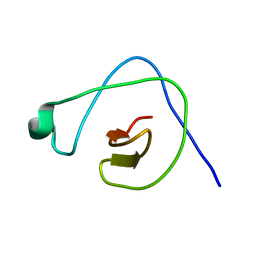 | | Solution structure of CpR82G from Clostridium perfringens. North East Structural Genomics Consortium Target CpR82g | | Descriptor: | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase domain protein, possible enterotoxin | | Authors: | Mobley, C.K, Lee, H, Lee, D, Ciccosanti, C, Janjua, H, Acton, T.B, Xiao, R, Everrett, J.K, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of CpR82G
To be Published
|
|
6K8W
 
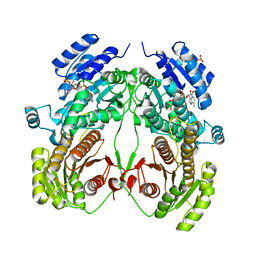 | | Crystal structure of N-domain with NADP of baterial malonyl-CoA reductase | | Descriptor: | NAD-dependent epimerase/dehydratase:Short-chain dehydrogenase/reductase SDR, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION | | Authors: | Kim, S, Kim, K.-J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Structural insight into bi-functional malonyl-CoA reductase.
Environ.Microbiol., 22, 2020
|
|
6KBV
 
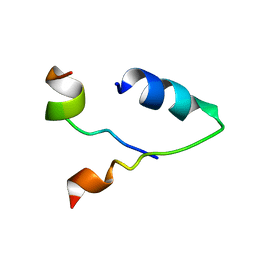 | |
6K8S
 
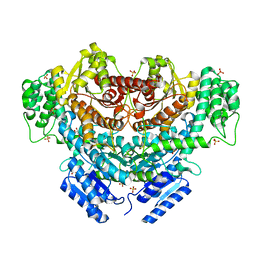 | |
6K8U
 
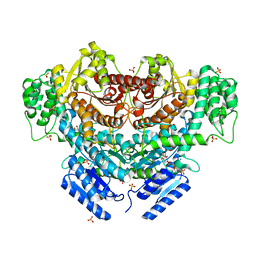 | |
6K8V
 
 | |
6KBO
 
 | | Three-dimensional LPS bound structure of VG16KRKP-KYE28. | | Descriptor: | Heparin cofactor 2, VG16KRKP | | Authors: | Ilyas, H, Bhunia, A. | | Deposit date: | 2019-06-26 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the combinatorial effects of antimicrobial peptides reveal a role of aromatic-aromatic interactions in antibacterial synergism.
J.Biol.Chem., 294, 2019
|
|
6K8T
 
 | |
4KSF
 
 | | Crystal Structure of Malonyl-CoA decarboxylase from Agrobacterium vitis, Northeast Structural Genomics Consortium Target RiR35 | | Descriptor: | CHLORIDE ION, Malonyl-CoA decarboxylase, NICKEL (II) ION | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of malonyl-coenzyme a decarboxylase provide insights into its catalytic mechanism and disease-causing mutations.
Structure, 21, 2013
|
|
2KT8
 
 | | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B | | Descriptor: | Probable surface protein | | Authors: | Yang, Y, Ramelot, T.A, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-01-21 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B
To be Published
|
|
7DZV
 
 | |
7DZT
 
 | | Cyrstal structure of PETase from Rhizobacter gummiphilus | | Descriptor: | DLH domain-containing protein | | Authors: | Sagong, H.-Y, Kim, K.-J. | | Deposit date: | 2021-01-26 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Implications for the PET decomposition mechanism through similarity and dissimilarity between PETases from Rhizobacter gummiphilus and Ideonella sakaiensis.
J Hazard Mater, 416, 2021
|
|
