1ZXE
 
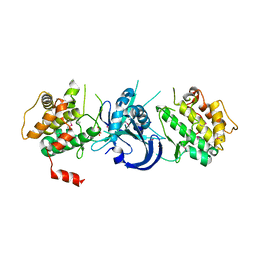 | | Crystal Structure of eIF2alpha Protein Kinase GCN2: D835N Inactivating Mutant in Apo Form | | Descriptor: | GLYCEROL, Serine/threonine-protein kinase | | Authors: | Padyana, A.K, Qiu, H, Roll-Mecak, A, Hinnebusch, A.G, Burley, S.K. | | Deposit date: | 2005-06-07 | | Release date: | 2005-06-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Basis for Autoinhibition and Mutational Activation of Eukaryotic Initiation Factor 2{alpha} Protein Kinase GCN2
J.Biol.Chem., 280, 2005
|
|
1UB4
 
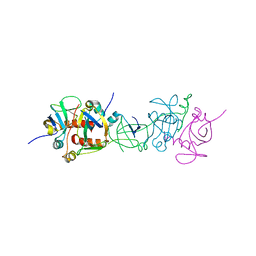 | |
3MZN
 
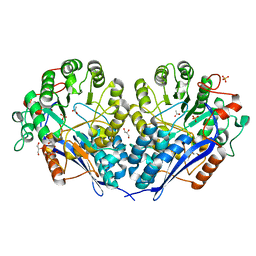 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 | | Descriptor: | ACETATE ION, GLYCEROL, Glucarate dehydratase, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-12 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
3N28
 
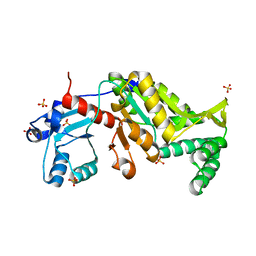 | | Crystal structure of probable phosphoserine phosphatase from vibrio cholerae, unliganded form | | Descriptor: | Phosphoserine phosphatase, SULFATE ION | | Authors: | Patskovsky, Y, Ramagopal, U, Toro, R, Rutter, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Phosphoserine Phosphatase from Vibrio Cholerae
To be Published
|
|
3N4E
 
 | | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans Pd1222 | | Descriptor: | CALCIUM ION, CHLORIDE ION, Mandelate racemase/muconate lactonizing enzyme, ... | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CRYSTAL STRUCTURE OF mandelate racemase/muconate lactonizing protein from Paracoccus denitrificans
Pd1222
To be Published
|
|
3N53
 
 | |
3N05
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM STREPTOMYCES AVERMITILIS | | Descriptor: | NH(3)-dependent NAD(+) synthetase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Do, J, Sauder, J.M, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of Nh3-Dependent Nad+ Synthetase from Streptomyces Avermitilis
To be Published
|
|
3N1U
 
 | | Structure of putative HAD superfamily (subfamily III A) hydrolase from Legionella pneumophila | | Descriptor: | CALCIUM ION, Hydrolase, HAD superfamily, ... | | Authors: | Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-07-21 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of putative HAD superfamily (subfamily III A) hydrolase from Legionella pneumophila
To be published
|
|
3MY9
 
 | | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans | | Descriptor: | GLYCEROL, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Quartararo, C.E, Ramagopal, U, Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-10 | | Release date: | 2010-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a muconate cycloisomerase from Azorhizobium caulinodans
To be Published
|
|
3N07
 
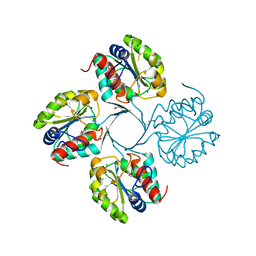 | | Structure of putative 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Vibrio cholerae | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, MAGNESIUM ION | | Authors: | Liu, W, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-13 | | Release date: | 2010-08-04 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis.
Biochemistry, 52, 2013
|
|
3NFU
 
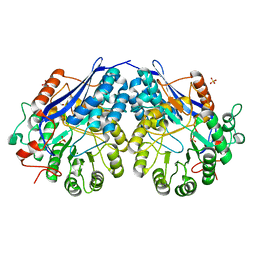 | | Crystal structure of probable glucarate dehydratase from chromohalobacter salexigens dsm 3043 complexed with magnesium | | Descriptor: | GLYCEROL, Glucarate dehydratase, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Sauder, J.M, Gerlt, J.A, Almo, S.C, Burley, S.K, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-10 | | Release date: | 2010-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal Structure of Glucarate Dehydratase from Chromohalobacter Salexigens
To be Published
|
|
3NHM
 
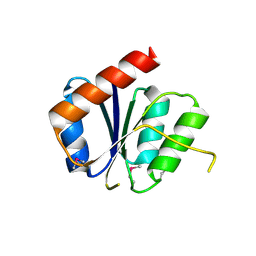 | |
3N2C
 
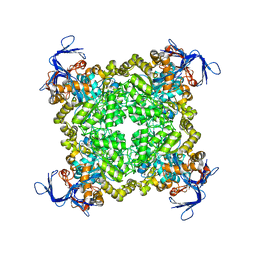 | | Crystal structure of prolidase eah89906 complexed with n-methylphosphonate-l-proline | | Descriptor: | 1-[(R)-hydroxy(methyl)phosphoryl]-L-proline, PROLIDASE, ZINC ION | | Authors: | Patskovsky, Y, Xu, C, Sauder, J.M, Burley, S.K, Raushel, F.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3N4F
 
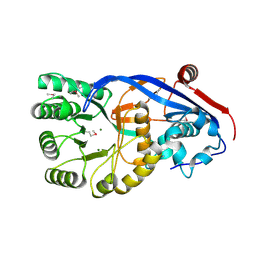 | | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing protein from Geobacillus sp. Y412MC10 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing protein from Geobacillus
sp. Y412MC10
To be Published
|
|
3NF5
 
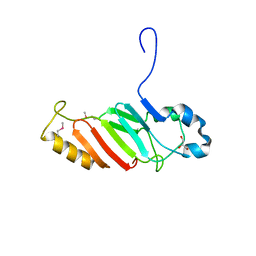 | | Crystal structure of the C-terminal domain of nuclear pore complex component NUP116 from Candida glabrata | | Descriptor: | GLYCEROL, Nucleoporin NUP116 | | Authors: | Sampathkumar, P, Manglicmot, D, Bain, K, Gilmore, J, Gheyi, T, Rout, M, Sali, A, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-08-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata.
Proteins, 80, 2012
|
|
3N3D
 
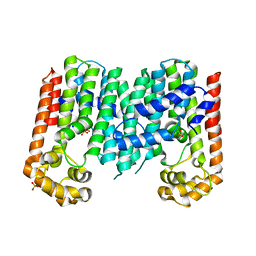 | | Crystal structure of geranylgeranyl pyrophosphate synthase from lactobacillus brevis atcc 367 | | Descriptor: | Geranylgeranyl pyrophosphate synthase, SULFATE ION | | Authors: | Patskovsky, Y, Toro, R, Rutter, M, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis
To be Published
|
|
3N6J
 
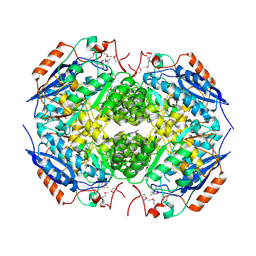 | | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus succinogenes 130Z | | Descriptor: | Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-25 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus
succinogenes 130Z
To be Published
|
|
3NEK
 
 | | Crystal structure of a nitrogen repressor-like protein MJ0159 from Methanococcus jannaschii | | Descriptor: | GLYCEROL, nitrogen repressor-like protein MJ0159 | | Authors: | Bonanno, J.B, Patskovsky, Y, Malashkevich, V, Ozyurt, S, Dickey, M, Wu, B, Maletic, M, Rodgers, L, Koss, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-09 | | Release date: | 2010-06-23 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural underpinnings of nitrogen regulation by the prototypical nitrogen-responsive transcriptional factor NrpR.
Structure, 18, 2010
|
|
3N6H
 
 | | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus succinogenes 130Z complexed with magnesium/sulfate | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-05-25 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mandelate racemase/muconate lactonizing protein from Actinobacillus
succinogenes 130Z complexed with magnesium/sulfate
To be Published
|
|
3NEH
 
 | | Crystal structure of the protein LMO2462 from Listeria monocytogenes complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala | | Descriptor: | (2R)-3-[(R)-[(1R)-1-amino-3-methylbutyl](hydroxy)phosphoryl]-2-methylpropanoic acid, Renal dipeptidase family protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Cummings, J, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-08 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of the protein LMO2462 from Listeria monocytogenes
complexed with ZN and phosphonate mimic of dipeptide L-Leu-D-Ala
To be Published
|
|
3NIW
 
 | | Crystal structure of a haloacid dehalogenase-like hydrolase from Bacteroides thetaiotaomicron | | Descriptor: | GLYCEROL, Haloacid dehalogenase-like hydrolase, MAGNESIUM ION | | Authors: | Bonanno, J.B, Ramagopal, U, Toro, R, Rutter, M, Bain, K.T, Wu, B, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2010-06-16 | | Release date: | 2010-06-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a haloacid dehalogenase-like hydrolase from Bacteroides thetaiotaomicron
To be Published
|
|
3NAS
 
 | |
3NIV
 
 | |
3NND
 
 | |
3NO1
 
 | |
