6ZTS
 
 | |
7RE9
 
 | | TCR mimic antibody (Fab fragment) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Fab heavy chain, Fab light chain | | Authors: | Dasgupta, M, Baker, B.M. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Validation and promise of a TCR mimic antibody for cancer immunotherapy of hepatocellular carcinoma.
Sci Rep, 12, 2022
|
|
7RE7
 
 | | TCR mimic antibody (Fab fragment) in complex with AFP/HLA-A*02 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, FORMIC ACID, ... | | Authors: | Dasgupta, M, Baker, B.M. | | Deposit date: | 2021-07-12 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Validation and promise of a TCR mimic antibody for cancer immunotherapy of hepatocellular carcinoma.
Sci Rep, 12, 2022
|
|
7RE8
 
 | |
1DK6
 
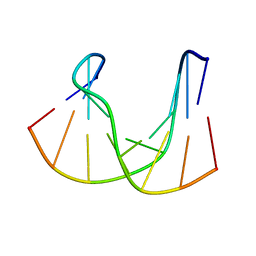 | | NMR structure analysis of the DNA nine base pair duplex D(CATGAGTAC) D(GTAC(NP3)CATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*(NP3)P*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 2000-01-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
1DK9
 
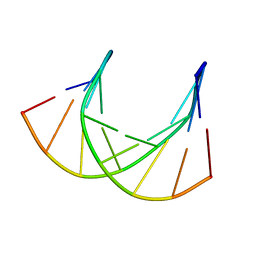 | | SOLUTION STRUCTURE ANALYSIS OF THE DNA DUPLEX D(CATGAGTAC)D(GTACTCATG) | | Descriptor: | 5'-D(CP*AP*TP*GP*AP*GP*TP*AP*CP*)-3', 5'-D(GP*TP*AP*CP*TP*CP*AP*TP*GP*)-3' | | Authors: | Klewer, D.A, Hoskins, A, Davisson, V.J, Bergstrom, D.E, LiWang, A.C. | | Deposit date: | 1999-12-06 | | Release date: | 1999-12-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a DNA duplex containing nucleoside analog 1-(2'-deoxy-beta-D-ribofuranosyl)-3-nitropyrrole and the structure of the unmodified control.
Nucleic Acids Res., 28, 2000
|
|
1KJU
 
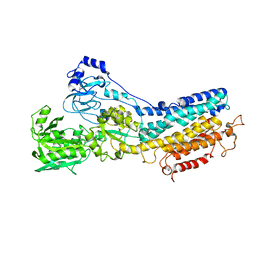 | | Ca2+-ATPase in the E2 State | | Descriptor: | Sarcoplasmic/endoplasmic reticulum calcium ATPase 1a | | Authors: | Xu, C, Rice, W.J, He, W, Stokes, D.L. | | Deposit date: | 2001-12-05 | | Release date: | 2001-12-19 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | A structural model for the catalytic cycle of Ca(2+)-ATPase.
J.Mol.Biol., 316, 2002
|
|
2PL5
 
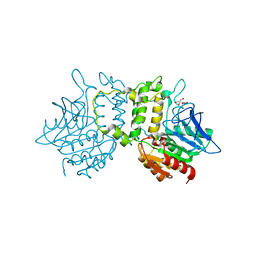 | | Crystal Structure of Homoserine O-acetyltransferase from Leptospira interrogans | | Descriptor: | GLYCEROL, Homoserine O-acetyltransferase | | Authors: | Liu, L, Wang, M, Wang, Y, Wei, Z, Xu, H, Gong, W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-11-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of homoserine O-acetyltransferase from Leptospira interrogans
Biochem.Biophys.Res.Commun., 363, 2007
|
|
3GST
 
 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | (9R,10R)-9-(S-GLUTATHIONYL)-10-HYDROXY-9,10-DIHYDROPHENANTHRENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Ammon, H.L, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
6DBL
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBR
 
 | | Cryo-EM structure of RAG in complex with one melted RSS and one unmelted RSS | | Descriptor: | CALCIUM ION, Forward strand of melted RSS substrate DNA, Forward strand of unmelted RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBQ
 
 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Molecule name: Forward strand of 12-RSS substrate DNA, Molecule name: Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.22 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBW
 
 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBU
 
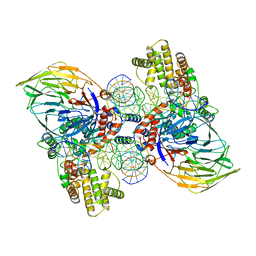 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBI
 
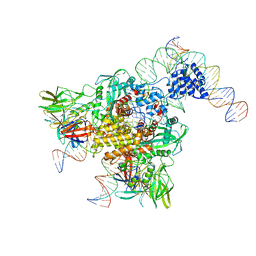 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS signal end, Forward strand of 23-RSS signal end, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBJ
 
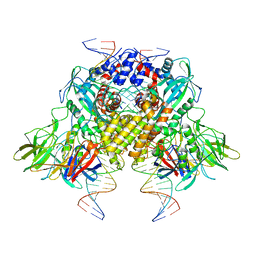 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS nicked DNA intermediates | | Descriptor: | CALCIUM ION, Forward stand of RSS signal end, Forward strand of coding flank, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DEF
 
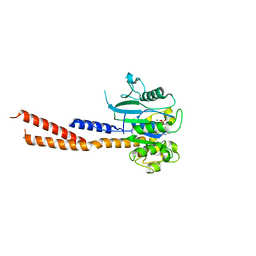 | | Vps1 GTPase-BSE fusion complexed with GMPPCP | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Vps1 GTPase-BSE | | Authors: | Ford, M.G.J, Varlakhanova, N.V, Brady, T.M, Chappie, J.S, Hosford, C.J. | | Deposit date: | 2018-05-11 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
6DI7
 
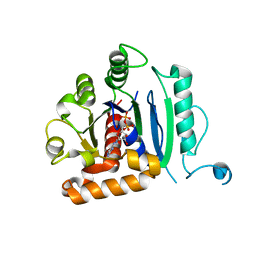 | | Vps1 GTPase-BSE fusion complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Putative sorting protein | | Authors: | Varlakhanova, N.V, Brady, T.M, Ford, M.G.J. | | Deposit date: | 2018-05-22 | | Release date: | 2018-08-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the fungal dynamin-related protein Vps1 reveal a unique, open helical architecture.
J. Cell Biol., 217, 2018
|
|
6DBT
 
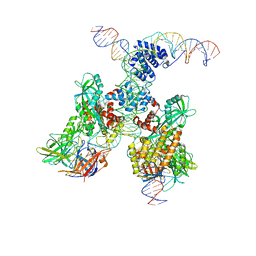 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6DBX
 
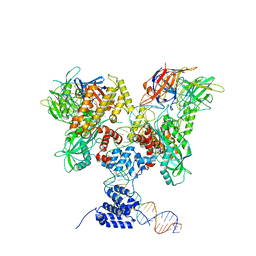 | | Cryo-EM structure of RAG in complex with 12-RSS substrate DNA | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Recombination activating gene 1 - MBP chimera, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6E0P
 
 | |
6E0C
 
 | |
6DZT
 
 | |
9JVM
 
 | |
9JVG
 
 | | Cryo-EM structure of the mmGPR4-Gs complex in pH6.2 | | Descriptor: | G-protein coupled receptor 4, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wen, X, Rong, N.K, Yang, F, Sun, J.P. | | Deposit date: | 2024-10-09 | | Release date: | 2025-02-26 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Evolutionary study and structural basis of proton sensing by Mus GPR4 and Xenopus GPR4.
Cell, 188, 2025
|
|
