7XVQ
 
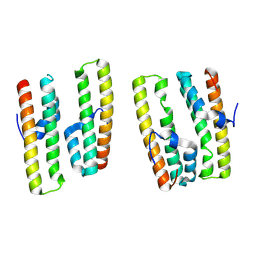 | | Crystal structure of AcrIIC4 | | Descriptor: | Uncharacterized protein | | Authors: | Zhang, H, Li, X.Z, Song, G.Y. | | Deposit date: | 2022-05-24 | | Release date: | 2023-08-09 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibitory mechanism of CRISPR-Cas9 by AcrIIC4.
Nucleic Acids Res., 51, 2023
|
|
8Z9C
 
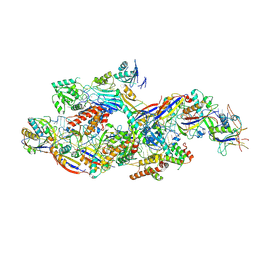 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | Protein structure, RNA (41-MER), RNA (48-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8YHE
 
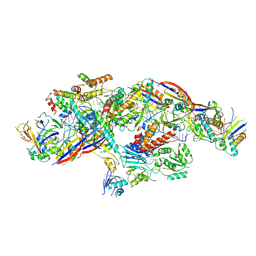 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state II | | Descriptor: | RNA (29-MER), RNA (46-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4J
 
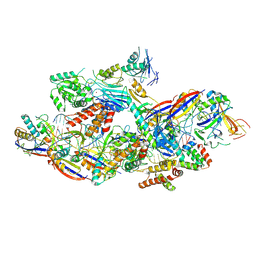 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state II | | Descriptor: | Protein structure, RNA (34-MER), RNA (38-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z4L
 
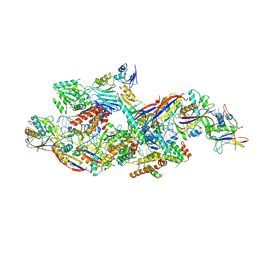 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at substrate-engaged state I | | Descriptor: | RNA (40-MER), RNA (49-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-17 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z9E
 
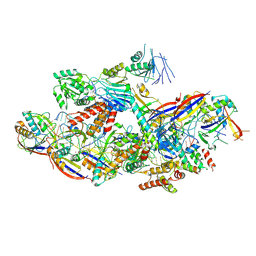 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state II | | Descriptor: | Protein structure, RNA (34-MER), RNA (39-MER), ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8Z99
 
 | | Cryo-EM structure of NTR-bound type VII CRISPR-Cas complex at substrate-engaged state +I | | Descriptor: | RNA (49-MER), RNA (54-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-04-22 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8YHD
 
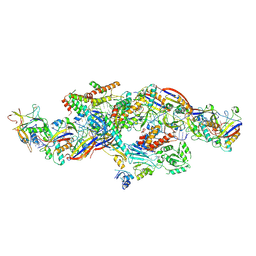 | | Cryo-EM structure of CTR-bound type VII CRISPR-Cas complex at post-state I | | Descriptor: | RNA (35-MER), RNA (53-MER), ZINC ION, ... | | Authors: | Zhang, H, Deng, Z, Li, X. | | Deposit date: | 2024-02-28 | | Release date: | 2024-08-21 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis for the activity of the type VII CRISPR-Cas system.
Nature, 633, 2024
|
|
8ZDY
 
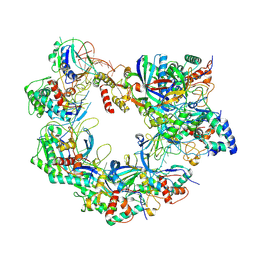 | | Cryo-EM structure of Cas8-HNH system at target free state | | Descriptor: | RNA (58-MER), a protein | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-05-03 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8ZNR
 
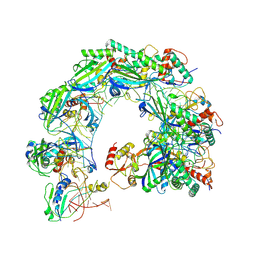 | | Cryo-EM structure of Cas8-HNH system at ssDNA-bound state | | Descriptor: | DNA (35-MER), RNA (56-MER), protein structure | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8IN8
 
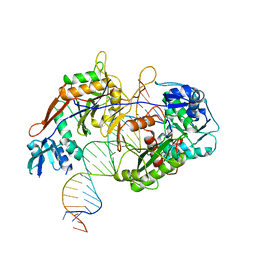 | | Cryo-EM structure of the target ssDNA-bound SIR2-APAZ/Ago-gRNA quaternary complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*GP*AP*CP*GP*TP*CP*TP*AP*AP*GP*AP*AP*AP*CP*CP*AP*TP*TP*AP*A)-3'), MAGNESIUM ION, Piwi domain protein, ... | | Authors: | Zhang, H, Li, Z, Yu, G.M, Li, X.Z, Wang, X.S. | | Deposit date: | 2023-03-08 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into mechanisms of Argonaute protein-associated NADase activation in bacterial immunity.
Cell Res., 33, 2023
|
|
8Z0L
 
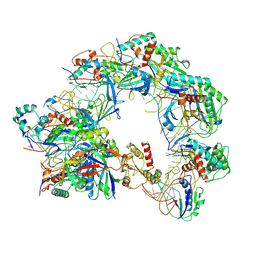 | | Cryo-EM structure of Cas8-HNH system at partial R-loop state | | Descriptor: | DNA (32-MER), DNA (5'-D(P*GP*TP*GP*CP*GP*GP*A)-3'), HNH endonuclease, ... | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-04-09 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8Z0K
 
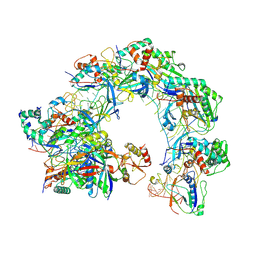 | | Cryo-EM structure of Cas8-HNH system at full R-loop state | | Descriptor: | DNA (37-MER), DNA (5'-D(P*GP*TP*GP*CP*GP*GP*A)-3'), HNH endonuclease, ... | | Authors: | Zhang, H, Zhu, H, Li, X, Liu, Y. | | Deposit date: | 2024-04-09 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural basis for the type I-F Cas8-HNH system.
Embo J., 43, 2024
|
|
8K98
 
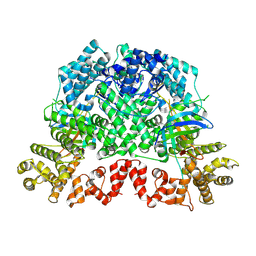 | | Cryo-EM structure of DSR2-TTP | | Descriptor: | a protein | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-01 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
8K9A
 
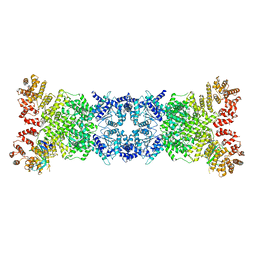 | | Cryo-EM structure of DSR2-DSAD1 state 2 | | Descriptor: | SIR2-like domain-containing protein, SPbeta prophage-derived uncharacterized protein YotI | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
7XND
 
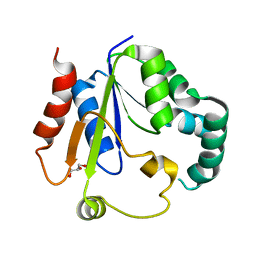 | | Crystal structure of Phosphomevalonate kinase from Silkworm | | Descriptor: | GLYCEROL, Phosphomevalonate kinase | | Authors: | Guo, P.C, Zhang, H. | | Deposit date: | 2022-04-28 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights into the substrate binding of phosphomevalonate kinase from the silkworm, Bombyx mori.
Insect Biochem.Mol.Biol., 150, 2022
|
|
9DYB
 
 | |
9DYA
 
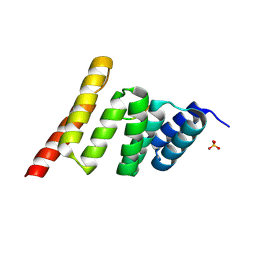 | | CHIP-TPR in complex with the Hsp70 tail | | Descriptor: | E3 ubiquitin-protein ligase CHIP, Heat shock 70 kDa protein 1A, SULFATE ION | | Authors: | Zhang, H, Nix, J.C, Page, R.C. | | Deposit date: | 2024-10-13 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Phosphorylation-State Modulated Binding of HSP70: Structural Insights and Compensatory Protein Engineering.
Biorxiv, 2025
|
|
5XAF
 
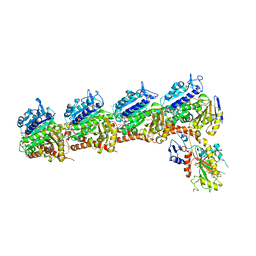 | | Crystal structure of tubulin-stathmin-TTL-Compound Z1 complex | | Descriptor: | (3S,4R)-4-(3-hydroxy-4-methoxyphenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhang, H, Luo, C, Wang, Y. | | Deposit date: | 2017-03-12 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Design, synthesis, biological evaluation and cocrystal structures with tubulin of chiral beta-lactam bridged combretastatin A-4 analogues as potent antitumor agents
Eur J Med Chem, 144, 2017
|
|
8FK2
 
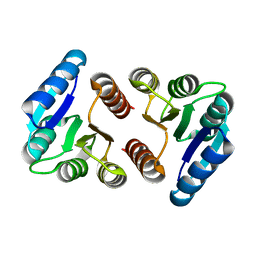 | | The N-terminal VicR from Streptococcus mutans | | Descriptor: | Putative response regulator CovR VicR-like protein | | Authors: | Zhang, H, Wu, H. | | Deposit date: | 2022-12-20 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Small Molecule Attenuates Bacterial Virulence by Targeting Conserved Response Regulator.
Mbio, 14, 2023
|
|
7WET
 
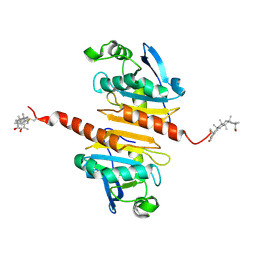 | | Crystal structure of Peroxiredoxin I in complex with the inhibitor Cela | | Descriptor: | (2R,4aS,6aS,12bR,14aS,14bR)-10-hydroxy-2,4a,6a,9,12b,14a-hexamethyl-11-oxo-1,2,3,4,4a,5,6,6a,11,12b,13,14,14a,14b-tetradecahydropicene-2-carboxylic acid, Peroxiredoxin-1 | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2021-12-24 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Celastrol suppresses colorectal cancer via covalent targeting peroxiredoxin 1.
Signal Transduct Target Ther, 8, 2023
|
|
7WEU
 
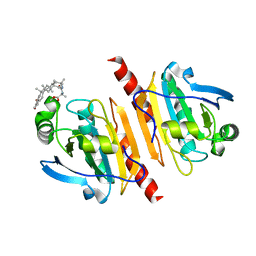 | |
8XKN
 
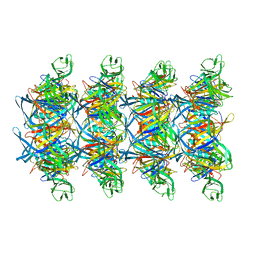 | | Cryo-EM structure of tail tube protein | | Descriptor: | a protein | | Authors: | Zhang, H, Li, Z, Li, X.Z. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Insights into the modulation of bacterial NADase activity by phage proteins.
Nat Commun, 15, 2024
|
|
6NMD
 
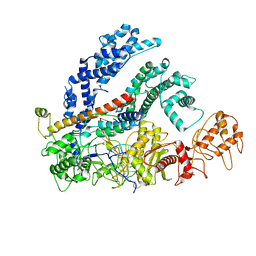 | | cryo-EM Structure of the LbCas12a-crRNA-AcrVA1 complex | | Descriptor: | AcrVA1, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
6NM9
 
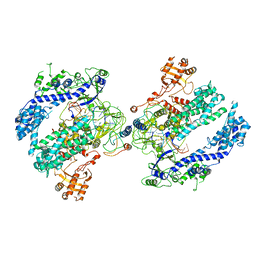 | | CryoEM structure of the LbCas12a-crRNA-AcrVA4 dimer | | Descriptor: | AcrVA4, Cpf1, MAGNESIUM ION, ... | | Authors: | Chang, L, Li, Z, Zhang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structural Basis for the Inhibition of CRISPR-Cas12a by Anti-CRISPR Proteins.
Cell Host Microbe, 25, 2019
|
|
