4TWY
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (S,R)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3S,4aR,8aS)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors
Bioorg.Med.Chem., 23, 2015
|
|
1VF9
 
 | | Solution Structure Of Human Trf2 | | Descriptor: | Telomeric repeat binding factor 2 | | Authors: | Nishimura, Y, Hanaoka, S. | | Deposit date: | 2004-04-12 | | Release date: | 2005-05-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Comparison between TRF2 and TRF1 of their telomeric DNA-bound structures and DNA-binding activities
Protein Sci., 14, 2005
|
|
6YYW
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 2-oxoglutarate, and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | 2-OXOGLUTARIC ACID, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase.
Chem Sci, 12, 2020
|
|
6YYY
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 4,4-dimethyl-2-oxoglutarate, and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | 2,2-dimethyl-4-oxidanylidene-pentanedioic acid, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase.
Chem Sci, 12, 2020
|
|
6Z6Q
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 3-ethyl-2-oxoglutarate, and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | (3~{R})-3-ethyl-2-oxidanylidene-pentanedioic acid, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-05-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase.
Chem Sci, 12, 2020
|
|
6Z6R
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, N-oxalyl-alpha-methylalanine, and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | Aspartyl/asparaginyl beta-hydroxylase, BROMIDE ION, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-05-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase.
Chem Sci, 12, 2020
|
|
6YYV
 
 | |
6YYU
 
 | |
6YYX
 
 | | Aspartyl/Asparaginyl beta-hydroxylase (AspH) oxygenase and TPR domains in complex with manganese, 3-methyl-2-oxoglutarate, and factor X substrate peptide fragment(39mer-4Ser) | | Descriptor: | (3~{R})-3-methyl-2-oxidanylidene-pentanedioic acid, Aspartyl/asparaginyl beta-hydroxylase, Coagulation factor X, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-05-06 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Synthesis of 2-oxoglutarate derivatives and their evaluation as cosubstrates and inhibitors of human aspartate/asparagine-beta-hydroxylase
Chem Sci, 12, 2021
|
|
7UXR
 
 | | Crystal structure of the BtTir TIR domain | | Descriptor: | TIR domain protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Vasquez, E, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXS
 
 | | Crystal structure of the BcThsA SLOG domain in complex with 3'cADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, BcThsA, GLYCEROL, ... | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXT
 
 | | Crystal structure of ligand-free SeThsA | | Descriptor: | GLYCEROL, TRIETHYLENE GLYCOL, USG protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Nanson, J.D, Kobe, B, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7XUA
 
 | | Structure of G9a in complex with compound 10a | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUD
 
 | | Structure of G9a in complex with compound 26a | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUC
 
 | | Structure of G9a in complex with compound 11a | | Descriptor: | 1,2-ETHANEDIOL, 3,6,6-trimethyl-4-oxidanylidene-~{N}-[(2~{S})-1-oxidanylidene-1-phenylazanyl-hexan-2-yl]-5,7-dihydro-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
7XUB
 
 | | Structure of G9a in complex with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Histone-lysine N-methyltransferase EHMT2, ... | | Authors: | Niwa, H, Shirai, F, Sato, S, Nishigaya, Y, Umehara, T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel Substrate-Competitive Lysine Methyltransferase G9a Inhibitors as Anticancer Agents.
J.Med.Chem., 66, 2023
|
|
4WY3
 
 | | Structure of SARS-3CL protease complex with a phenylbenzoyl (R,S)-N-decalin type inhibitor | | Descriptor: | (2S)-2-({[(3R,4aS,8aR)-2-(biphenyl-4-ylcarbonyl)decahydroisoquinolin-3-yl]methyl}amino)-3-(1H-imidazol-5-yl)propanal, 3C-like proteinase | | Authors: | Akaji, K, Teruya, K, Shimamoto, Y, Sanjho, A, Yamashita, E, Nakagawa, A. | | Deposit date: | 2014-11-15 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Fused-ring structure of decahydroisoquinolin as a novel scaffold for SARS 3CL protease inhibitors.
Bioorg.Med.Chem., 23, 2015
|
|
3VWL
 
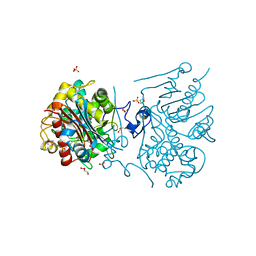 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187S/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
8KI1
 
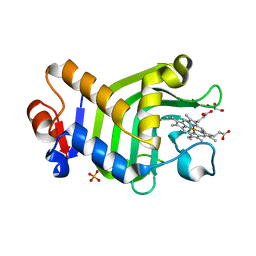 | | Crystal structure of the holo form of the hemophore HasA from Pseudomonas protegens Pf-5 | | Descriptor: | GLYCEROL, Heme acquisition protein HasAp, PHOSPHATE ION, ... | | Authors: | Shisaka, Y, Inaba, H, Sugimoto, H, Shoji, O. | | Deposit date: | 2023-08-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Heme-substituted protein assembly bridged by synthetic porphyrin: achieving controlled configuration while maintaining rotational freedom.
Rsc Adv, 14, 2024
|
|
8KI0
 
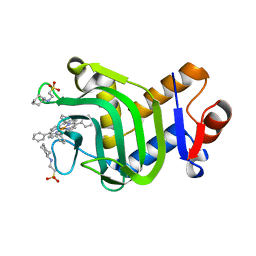 | | Crystal structure of the hemophore HasA from Pseudomonas protegens Pf-5 capturing Fe-tetraphenylporphyrin | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Heme acquisition protein HasAp, ... | | Authors: | Shisaka, Y, Inaba, H, Sugimoto, H, Shoji, O. | | Deposit date: | 2023-08-22 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Heme-substituted protein assembly bridged by synthetic porphyrin: achieving controlled configuration while maintaining rotational freedom.
Rsc Adv, 14, 2024
|
|
3VWR
 
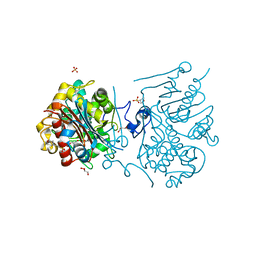 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187G/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
5AVE
 
 | | The ligand binding domain of Mlp37 with serine | | Descriptor: | Methyl-accepting chemotaxis (MCP) signaling domain protein, SERINE | | Authors: | Takahashi, Y, Sumita, K, Uchida, Y, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2015-06-15 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants
Sci Rep, 6, 2016
|
|
3VWM
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase G181D/R187A/H266N/D370Y mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
To be Published
|
|
3VWQ
 
 | | 6-aminohexanoate-dimer hydrolase S112A/G181D/R187A/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
3VWP
 
 | | Crystal structure of 6-aminohexanoate-dimer hydrolase S112A/G181D/R187S/H266N/D370Y mutant complexd with 6-aminohexanoate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Kawashima, Y, Shibata, N, Negoro, S, Higuchi, Y. | | Deposit date: | 2012-08-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural, kinetic and theoretical analyses of hydrolase mutants altering in the directionality and equilibrium point of reversible amide-synthetic/hydrolytic reaction
to be published
|
|
