1RHI
 
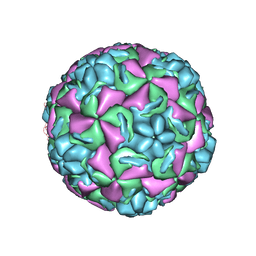 | | HUMAN RHINOVIRUS 3 COAT PROTEIN | | Descriptor: | CALCIUM ION, HUMAN RHINOVIRUS 3 COAT PROTEIN | | Authors: | Zhao, R, Rossmann, M.G. | | Deposit date: | 1996-06-17 | | Release date: | 1997-03-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Human rhinovirus 3 at 3.0 A resolution.
Structure, 4, 1996
|
|
1RVF
 
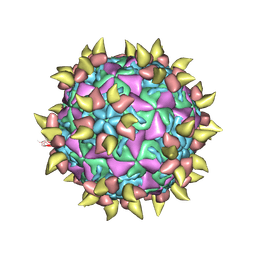 | | FAB COMPLEXED WITH INTACT HUMAN RHINOVIRUS | | Descriptor: | FAB 17-IA, HUMAN RHINOVIRUS 14 COAT PROTEIN | | Authors: | Smith, T.J. | | Deposit date: | 1996-09-05 | | Release date: | 1998-02-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon.
Nature, 383, 1996
|
|
1VRH
 
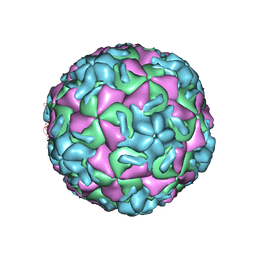 | | HRV14/SDZ 880-061 COMPLEX | | Descriptor: | 2-[4-(2H-1,4-BENZOTHIAZINE-3-YL)-PIPERAZINE-1-LY]-1,3-THIAZOLE-4-CARBOXYLIC ACID ETHYLESTER, RHINOVIRUS 14 | | Authors: | Oren, D.A, Zhang, A, Arnold, E. | | Deposit date: | 1996-02-26 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and activity of piperazine-containing antirhinoviral agents and crystal structure of SDZ 880-061 bound to human rhinovirus 14.
J.Mol.Biol., 259, 1996
|
|
1D3I
 
 | |
2O3C
 
 | | Crystal structure of zebrafish Ape | | Descriptor: | APEX nuclease 1, LEAD (II) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
2O3H
 
 | | Crystal structure of the human C65A Ape | | Descriptor: | ACETATE ION, DNA-(apurinic or apyrimidinic site) lyase, SAMARIUM (III) ION | | Authors: | Georgiadis, M.M, Gaur, R.K, Delaplane, S, Svenson, J. | | Deposit date: | 2006-12-01 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Evolution of the redox function in mammalian apurinic/apyrimidinic endonuclease
Mutat.Res., 643, 2008
|
|
5W1Y
 
 | | SETD8 in complex with a covalent inhibitor | | Descriptor: | 2-(4-methylpiperazin-1-yl)-3-(phenylsulfanyl)naphthalene-1,4-dione, N-lysine methyltransferase KMT5A, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Yu, W, Li, Y, Blum, G, Luo, M, Pittella-Silva, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-05 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | SETD8 in complex with a covalent inhibitor
to be published
|
|
