4YNU
 
 | | Crystal structure of Aspergillus flavus FADGDH in complex with D-glucono-1,5-lactone | | Descriptor: | D-glucono-1,5-lactone, FLAVIN-ADENINE DINUCLEOTIDE, Glucose oxidase, ... | | Authors: | Yoshida, H, Sakai, G, Kojima, K, Kamitori, S, Sode, K. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural analysis of fungus-derived FAD glucose dehydrogenase
Sci Rep, 5, 2015
|
|
1CTO
 
 | | NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF THE LIGAND-BINDING REGION OF MURINE GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR | | Authors: | Yamasaki, K, Naito, S, Anaguchi, H, Ohkubo, T, Ota, Y. | | Deposit date: | 1996-09-25 | | Release date: | 1997-10-22 | | Last modified: | 2018-03-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an extracellular domain containing the WSxWS motif of the granulocyte colony-stimulating factor receptor and its interaction with ligand.
Nat.Struct.Biol., 4, 1997
|
|
4YNT
 
 | | Crystal structure of Aspergillus flavus FAD glucose dehydrogenase | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Glucose oxidase, putative | | Authors: | Yoshida, H, Sakai, G, Kojima, K, Kamitori, S, Sode, K. | | Deposit date: | 2015-03-11 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural analysis of fungus-derived FAD glucose dehydrogenase
Sci Rep, 5, 2015
|
|
4XSM
 
 | | Crystal structure of D-tagatose 3-epimerase C66S from Pseudomonas cichorii in complex with D-talitol | | Descriptor: | D-altritol, D-tagatose 3-epimerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yoshihara, A, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structures of the Pseudomonas cichorii D-tagatose 3-epimerase mutant form C66S recognizing deoxy sugars as substrates
Appl. Microbiol. Biotechnol., 100, 2016
|
|
3NV1
 
 | | Crystal structure of human galectin-9 C-terminal CRD | | Descriptor: | Galectin 9 short isoform variant, NICKEL (II) ION | | Authors: | Yoshida, H, Kamitori, S. | | Deposit date: | 2010-07-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray structures of human galectin-9 C-terminal domain in complexes with a biantennary oligosaccharide and sialyllactose
J.Biol.Chem., 285, 2010
|
|
7CJ4
 
 | |
7CJ9
 
 | | Crystal structure of N-terminal His-tagged D-allulose 3-epimerase from Methylomonas sp. with additional C-terminal residues | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-fructose, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ7
 
 | | Crystal structure of homo dimeric D-allulose 3-epimerase from Methylomonas sp. in complex with L-tagatose | | Descriptor: | (2R,3R,4R,5S)-2-(hydroxymethyl)oxane-2,3,4,5-tetrol, Epimerase, L-sorbose, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ6
 
 | |
7CJ8
 
 | | Crystal structure of N-terminal His-tagged D-allulose 3-epimerase from Methylomonas sp. in complex with D-allulose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, D-psicose, Epimerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
7CJ5
 
 | | Crystal structure of homo dimeric D-allulose 3-epimerase from Methylomonas sp. in complex with D-fructose | | Descriptor: | D-fructose, Epimerase, MAGNESIUM ION, ... | | Authors: | Yoshida, H, Yoshihara, A, Kamitori, S. | | Deposit date: | 2020-07-09 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel homodimeric l-ribulose 3-epimerase from Methylomonus sp.
Febs Open Bio, 11, 2021
|
|
1JF6
 
 | | Crystal structure of thermoactinomyces vulgaris r-47 alpha-amylase mutant F286Y | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
4FQZ
 
 | |
1JF5
 
 | | CRYSTAL STRUCTURE OF THERMOACTINOMYCES VULGARIS R-47 ALPHA-AMYLASE 2 MUTANT F286A | | Descriptor: | ALPHA AMYLASE II, CALCIUM ION | | Authors: | Ohtaki, A, Kondo, S, Shimura, Y, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2001-06-20 | | Release date: | 2002-05-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Role of Phe286 in the recognition mechanism of cyclomaltooligosaccharides (cyclodextrins) by Thermoactinomyces vulgaris R-47 alpha-amylase 2 (TVAII). X-ray structures of the mutant TVAIIs, F286A and F286Y, and kinetic analyses of the Phe286-replaced mutant TVAIIs
CARBOHYDR.RES., 334, 2001
|
|
5T1F
 
 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase mutant Asn56Ala | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Uncharacterized protein | | Authors: | Yoshida, H, Shimasaki, T, Kamitori, S, Sode, K. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
5T1E
 
 | | Crystal structure of Phaeospaeria nodrum fructosyl peptide oxidase | | Descriptor: | ACETIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Uncharacterized protein | | Authors: | Yoshida, H, Shimasaki, T, Kamitori, S, Sode, K. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | X-ray structures of fructosyl peptide oxidases revealing residues responsible for gating oxygen access in the oxidative half reaction
Sci Rep, 7, 2017
|
|
3ITY
 
 | | Metal-free form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITO
 
 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant D327N in complex with D-psicose | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION, alpha-D-psicofuranose | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITT
 
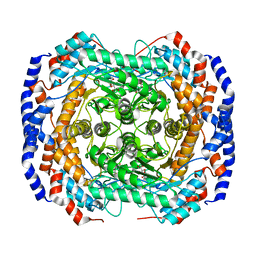 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329K in complex with L-rhamnose | | Descriptor: | L-RHAMNOSE, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUD
 
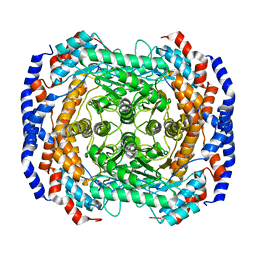 | | Cu2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COPPER (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITL
 
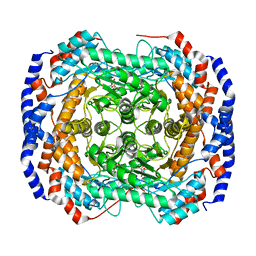 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant D327N in complex with L-rhamnulose | | Descriptor: | 6-deoxy-beta-L-fructofuranose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUI
 
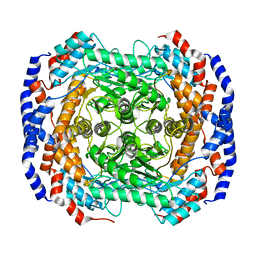 | | Zn2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITX
 
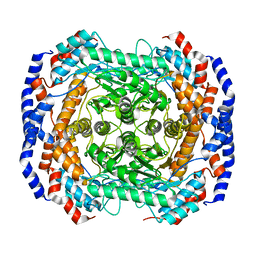 | | Mn2+ bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3ITV
 
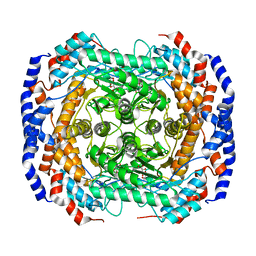 | | Crystal structure of Pseudomonas stutzeri L-rhamnose isomerase mutant S329K in complex with D-psicose | | Descriptor: | D-psicose, L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
3IUH
 
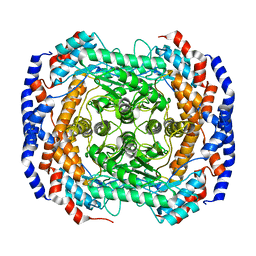 | | Co2+-bound form of Pseudomonas stutzeri L-rhamnose isomerase | | Descriptor: | COBALT (II) ION, L-rhamnose isomerase | | Authors: | Yoshida, H, Yamaji, M, Ishii, T, Izumori, K, Kamitori, S. | | Deposit date: | 2009-08-31 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic reaction mechanism of Pseudomonas stutzeri l-rhamnose isomerase deduced from X-ray structures
Febs J., 277, 2010
|
|
