6LZG
 
 | | Structure of novel coronavirus spike receptor-binding domain complexed with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Wang, Q.H, Song, H, Qi, J.X. | | Deposit date: | 2020-02-19 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Basis of SARS-CoV-2 Entry by Using Human ACE2.
Cell, 181, 2020
|
|
2VR5
 
 | | Crystal structure of Trex from Sulfolobus Solfataricus in complex with acarbose intermediate and glucose | | Descriptor: | 4-O-(4,6-dideoxy-4-{[(1S,2S,3S,4R,5S)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranosyl)-beta-D-glucopyranose, GLYCEROL, GLYCOGEN OPERON PROTEIN GLGX, ... | | Authors: | Song, H.-N, Yoon, S.-M, Lee, S.-J, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-03-26 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
2VUY
 
 | | Crystal structure of Glycogen Debranching exzyme TreX from Sulfolobus solfatarius | | Descriptor: | GLYCOGEN OPERON PROTEIN GLGX | | Authors: | Song, H.-N, Yoon, S.-M, Cha, H.-J, Park, K.-H, Woo, E.-J. | | Deposit date: | 2008-06-02 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Insight Into the Bifunctional Mechanism of the Glycogen-Debranching Enzyme Trex from the Archaeon Sulfolobus Solfataricus.
J.Biol.Chem., 283, 2008
|
|
6EF4
 
 | | Crystal structure of mouse PP2A Aalpha P179R mutant | | Descriptor: | Serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform | | Authors: | Wang, Z, Shen, G, Xu, W. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Highly Recurrent PP2A A alpha-Subunit Mutation P179R Alters Protein Structure and Impairs PP2A Enzyme Function to Promote Endometrial Tumorigenesis.
Cancer Res., 79, 2019
|
|
4XG2
 
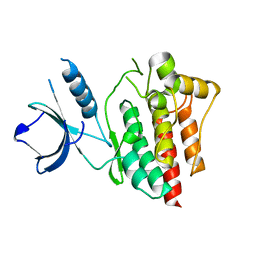 | | Crystal structure of ligand-free Syk | | Descriptor: | Tyrosine-protein kinase SYK | | Authors: | Lee, S.J, Choi, J, Han, B.G, Song, H, Koh, J.S, Lee, B.I. | | Deposit date: | 2014-12-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structures of spleen tyrosine kinase in complex with novel inhibitors: structural insights for design of anticancer drugs
Febs J., 283, 2016
|
|
6KUT
 
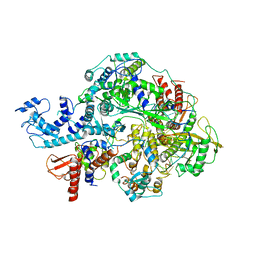 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KV5
 
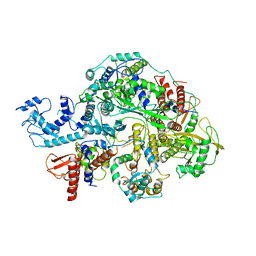 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
7M72
 
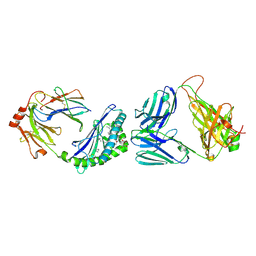 | | MHC-like protein complex structure | | Descriptor: | (3R)-N-[(2S,3R)-1-(alpha-D-galactopyranosyloxy)-3-hydroxy-15-methylhexadecan-2-yl]-3-hydroxyheptadecanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Thirunavukkarasu, P, Le Nours, J, Rossjohn, J. | | Deposit date: | 2021-03-26 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Host immunomodulatory lipids created by symbionts from dietary amino acids.
Nature, 600, 2021
|
|
3ZHL
 
 | |
3ZHK
 
 | |
3ZHD
 
 | |
2H5E
 
 | | Crystal structure of E.coli polypeptide release factor RF3 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Peptide chain release factor RF-3 | | Authors: | Song, H.W, Zhou, Z.H. | | Deposit date: | 2006-05-26 | | Release date: | 2007-05-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors
Cell(Cambridge,Mass.), 129, 2007
|
|
4Y5T
 
 | | Structure of FtmOx1 apo with metal Iron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COBALT (II) ION, FE (II) ION, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
4Y5S
 
 | | Structure of FtmOx1 with a-Ketoglutarate as co-substrate | | Descriptor: | 2-OXOGLUTARIC ACID, COBALT (II) ION, FE (II) ION, ... | | Authors: | Yan, W, Zhang, Y. | | Deposit date: | 2015-02-12 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Endoperoxide formation by an alpha-ketoglutarate-dependent mononuclear non-haem iron enzyme.
Nature, 527, 2015
|
|
6J6M
 
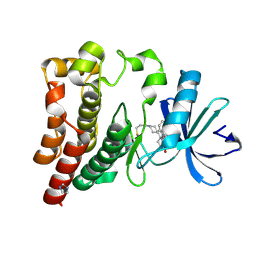 | | Co-crystal structure of BTK kinase domain with Zanubrutinib | | Descriptor: | (7S)-2-(4-phenoxyphenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, IMIDAZOLE, Tyrosine-protein kinase BTK | | Authors: | Zhou, X, Hong, Y. | | Deposit date: | 2019-01-15 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of Zanubrutinib (BGB-3111), a Novel, Potent, and Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 62, 2019
|
|
7BRP
 
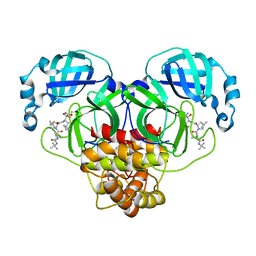 | |
7BRO
 
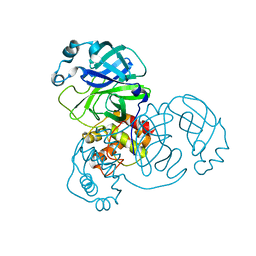 | | Crystal structure of the 2019-nCoV main protease | | Descriptor: | 3C-like proteinase | | Authors: | Fu, L.F. | | Deposit date: | 2020-03-29 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease.
Nat Commun, 11, 2020
|
|
6LP5
 
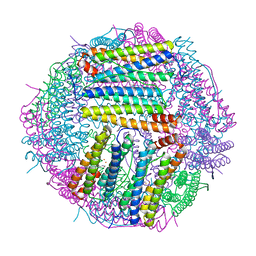 | | Structure of Sinonovacula constricta ferritin | | Descriptor: | FE (II) ION, FE (III) ION, Ferritin, ... | | Authors: | Su, X.R, Ming, T.H, Su, C. | | Deposit date: | 2020-01-08 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystallographic characterization of ferritin from Sinonovacula constricta.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
4I4Z
 
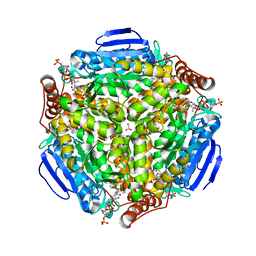 | | Synechocystis sp. PCC 6803 1,4-dihydroxy-2-naphthoyl-coenzyme A synthase (MenB) in complex with salicylyl-CoA | | Descriptor: | BICARBONATE ION, MALONATE ION, Naphthoate synthase, ... | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
4I52
 
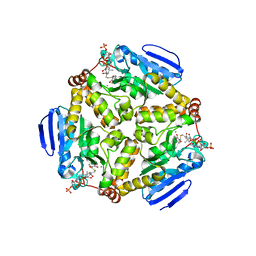 | | scMenB im complex with 1-hydroxy-2-naphthoyl-CoA | | Descriptor: | 1-hydroxy-2-naphthoyl-CoA, CHLORIDE ION, Naphthoate synthase | | Authors: | Song, H.G, Sun, Y.R, Li, J, Li, Y, Jiang, M, Zhou, J.H, Guo, Z.H. | | Deposit date: | 2012-11-28 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the induced-fit mechanism of 1,4-dihydroxy-2-naphthoyl coenzyme a synthase from the crotonase fold superfamily
Plos One, 8, 2013
|
|
6KUJ
 
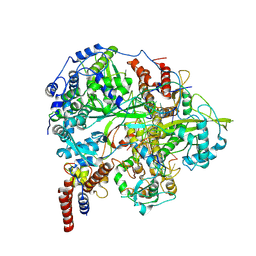 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 1 | | Descriptor: | 3'-cRNA promoter, 5'-cRNA promoter, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of influenza D virus polymerase bound to cRNA promoter in Mode A conformation
NAT NANOTECHNOL, 2019
|
|
2GIZ
 
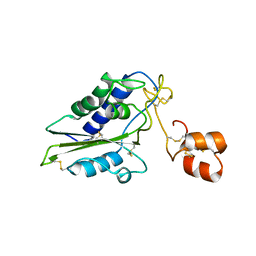 | | Structural and functional analysis of Natrin, a member of crisp-3 family blocks a variety of ion channels | | Descriptor: | Natrin-1 | | Authors: | Jiang, T, Wang, F, Li, H, Yin, C, Zhou, Y, Shu, Y, Qi, Z, Lin, Z. | | Deposit date: | 2006-03-30 | | Release date: | 2006-11-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and functional analysis of natrin, a venom protein that targets various ion channels
Biochem.Biophys.Res.Commun., 351, 2006
|
|
5XVF
 
 | | Crystal Structure of PAK4 in complex with inhibitor CZH062 | | Descriptor: | 2-(4-azanylpiperidin-1-yl)-6-chloranyl-N-(1-methylimidazol-4-yl)quinazolin-4-amine, Serine/threonine-protein kinase PAK 4 | | Authors: | Zhao, F, Li, H. | | Deposit date: | 2017-06-27 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.655 Å) | | Cite: | Structure-Based Design of 6-Chloro-4-aminoquinazoline-2-carboxamide Derivatives as Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors.
J. Med. Chem., 61, 2018
|
|
3EIJ
 
 | | Crystal structure of Pdcd4 | | Descriptor: | Programmed cell death protein 4 | | Authors: | Loh, P.G. | | Deposit date: | 2008-09-16 | | Release date: | 2009-02-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for translational inhibition by the tumour suppressor Pdcd4
Embo J., 28, 2009
|
|
3D3J
 
 | | Crystal structure of human Edc3p | | Descriptor: | Enhancer of mRNA-decapping protein 3 | | Authors: | Ling, S.H.M. | | Deposit date: | 2008-05-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human Edc3 and its functional implications
Mol.Cell.Biol., 28, 2008
|
|
