3AFP
 
 | | Crystal structure of the single-stranded DNA binding protein from Mycobacterium leprae (Form I) | | Descriptor: | CADMIUM ION, GLYCEROL, Single-stranded DNA-binding protein | | Authors: | Kaushal, P.S, Singh, P, Sharma, A, Muniyappa, K, Vijayan, M. | | Deposit date: | 2010-03-10 | | Release date: | 2010-10-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray and molecular-dynamics studies on Mycobacterium leprae single-stranded DNA-binding protein and comparison with other eubacterial SSB structures
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3KK2
 
 | | HIV-1 reverse transcriptase-DNA complex with dATP bound in the nucleotide binding site | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*A*TP*GP*GP*TP*GP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3', 5'-D(*AP*CP*A*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DOC))-3', ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2009-11-04 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Visualizing the molecular interactions of a nucleotide analog, GS-9148, with HIV-1 reverse transcriptase-DNA complex.
J.Mol.Biol., 397, 2010
|
|
5W6Z
 
 | | Crystal structure of the H24W mutant of HsNUDT16 | | Descriptor: | SODIUM ION, U8 snoRNA-decapping enzyme | | Authors: | Thirawatananond, P, Gabelli, S.B. | | Deposit date: | 2017-06-18 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural analyses of NudT16-ADP-ribose complexes direct rational design of mutants with improved processing of poly(ADP-ribosyl)ated proteins.
Sci Rep, 9, 2019
|
|
1BI2
 
 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR | | Authors: | Pohl, E, Hol, W.G.J. | | Deposit date: | 1998-06-21 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
3FBR
 
 | | structure of HipA-amppnp-peptide | | Descriptor: | PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Serine/threonine-protein kinase toxin HipA, peptide of EF-Tu | | Authors: | Schumacher, M.A. | | Deposit date: | 2008-11-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
5W6X
 
 | |
1BI0
 
 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR, SULFATE ION, ZINC ION | | Authors: | Pohl, E, Hol, W.G. | | Deposit date: | 1998-06-21 | | Release date: | 1999-07-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
1BI1
 
 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR | | Authors: | Pohl, E, Hol, W.G.J. | | Deposit date: | 1998-06-21 | | Release date: | 1999-06-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
1BI3
 
 | | STRUCTURE OF APO-AND HOLO-DIPHTHERIA TOXIN REPRESSOR | | Descriptor: | DIPHTHERIA TOXIN REPRESSOR, SULFATE ION, ZINC ION | | Authors: | Pohl, E, Hol, W.G.J. | | Deposit date: | 1998-06-21 | | Release date: | 1999-06-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Motion of the DNA-binding domain with respect to the core of the diphtheria toxin repressor (DtxR) revealed in the crystal structures of apo- and holo-DtxR.
J.Biol.Chem., 273, 1998
|
|
5W2J
 
 | | Crystal structure of dimeric form of mouse Glutaminase C | | Descriptor: | CHLORIDE ION, Glutaminase kidney isoform, mitochondrial, ... | | Authors: | Cerione, R.A, Li, Y. | | Deposit date: | 2017-06-06 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanistic Basis of Glutaminase Activation: A KEY ENZYME THAT PROMOTES GLUTAMINE METABOLISM IN CANCER CELLS.
J. Biol. Chem., 291, 2016
|
|
3GTU
 
 | |
3LPP
 
 | | Crystal complex of N-terminal sucrase-isomaltase with kotalanol | | Descriptor: | (1S,2R,3R,4S)-1-{(1S)-2-[(2R,3S,4S)-3,4-dihydroxy-2-(hydroxymethyl)tetrahydrothiophenium-1-yl]-1-hydroxyethyl}-2,3,4,5-tetrahydroxypentyl sulfate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains.
J.Biol.Chem., 285, 2010
|
|
2OKT
 
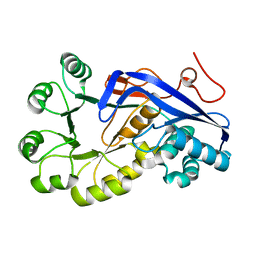 | | Crystal structure of O-succinylbenzoic acid synthetase from Staphylococcus aureus, ligand-free form | | Descriptor: | O-succinylbenzoic acid synthetase | | Authors: | Patskovsky, Y, Toro, R, Malashkevich, V, Sauder, J.M, Ozyurt, S, Smith, D, Dickey, M, Maletic, M, Powell, A, Gheyi, T, Wasserman, S.R, Gerlt, J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-01-17 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5IKK
 
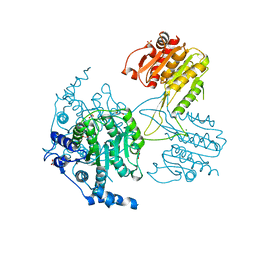 | | Structure of the histone deacetylase Clr3 | | Descriptor: | 1,2-ETHANEDIOL, Histone deacetylase clr3, MAGNESIUM ION, ... | | Authors: | Brugger, C, Schalch, T. | | Deposit date: | 2016-03-03 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SHREC Silences Heterochromatin via Distinct Remodeling and Deacetylation Modules.
Mol.Cell, 62, 2016
|
|
5IQY
 
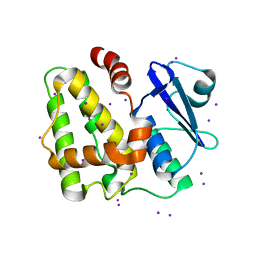 | | Structure of apo-Dehydroascorbate Reductase from Pennisetum Glaucum phased by Iodide-SAD method | | Descriptor: | Dehydroascorbate reductase, IODIDE ION | | Authors: | Das, B.K, Kumar, A, Manidola, P, Arockiasamy, A. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Non-native ligands define the active site of Pennisetum glaucum (L.) R. Br dehydroascorbate reductase
Biochem.Biophys.Res.Commun., 473, 2016
|
|
5IKF
 
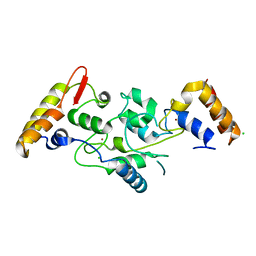 | |
3C6B
 
 | |
3LPO
 
 | | Crystal structure of the N-terminal domain of sucrase-isomaltase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Sucrase-isomaltase, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2010-02-05 | | Release date: | 2010-03-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for substrate selectivity in human maltase-glucoamylase and sucrase-isomaltase N-terminal domains.
J.Biol.Chem., 285, 2010
|
|
2OZT
 
 | | Crystal structure of O-succinylbenzoate synthase from Thermosynechococcus elongatus BP-1 | | Descriptor: | PHOSPHATE ION, SODIUM ION, Tlr1174 protein | | Authors: | Malashkevich, V.N, Bonanno, J, Toro, R, Sauder, J.M, Schwinn, K.D, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Gheyi, T, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-02-27 | | Release date: | 2007-03-13 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Loss of quaternary structure is associated with rapid sequence divergence in the OSBS family.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2IVV
 
 | | Crystal structure of phosphorylated RET tyrosine kinase domain complexed with the inhibitor PP1 | | Descriptor: | 1-TER-BUTYL-3-P-TOLYL-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4-YLAMINE, FORMIC ACID, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE RECEPTOR RET PRECURSOR | | Authors: | Knowles, P.P, Murray-Rust, J, McDonald, N.Q. | | Deposit date: | 2006-06-16 | | Release date: | 2006-08-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and chemical inhibition of the RET tyrosine kinase domain.
J. Biol. Chem., 281, 2006
|
|
2IVU
 
 | |
7X27
 
 | | MERS-CoV spike complex | | Descriptor: | Spike glycoprotein | | Authors: | Zeng, J.W, Zhang, S.Y, Wang, X.W. | | Deposit date: | 2022-02-25 | | Release date: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | cryo-EM structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein
To Be Published
|
|
5XFI
 
 | |
5XFH
 
 | | Crystal structure of Orysata lectin in complex with biantennary N-glycan | | Descriptor: | Salt stress-induced protein, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose | | Authors: | Nagae, M, Yamaguchi, Y. | | Deposit date: | 2017-04-10 | | Release date: | 2017-10-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Distinct roles for each N-glycan branch interacting with mannose-binding type Jacalin-related lectins Orysata and Calsepa.
Glycobiology, 27, 2017
|
|
1D88
 
 | |
