5R5P
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13910a | | Descriptor: | DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22, ~{N}-[(4-fluorophenyl)methyl]-4-methoxy-aniline | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R69
 
 | | XChem group deposition -- Crystal Structure of human YEATS4 in complex with XS038644e | | Descriptor: | 1,2-ETHANEDIOL, YEATS domain-containing protein 4, ~{N}-(5-oxidanylidene-7,8-dihydro-6~{H}-naphthalen-2-yl)ethanamide | | Authors: | Raux, B, Krojer, T, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Huber, K.V.M. | | Deposit date: | 2020-02-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5RF0
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with POB0073 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, [1-(pyridin-2-yl)cyclopentyl]methanol | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFI
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102353 | | Descriptor: | 1-{4-[(2,5-dimethylphenyl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFW
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102243 | | Descriptor: | 1-{4-[(thiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RGU
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z4444622180 (Mpro-x2562) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(3-{[(2R)-4-oxoazetidin-2-yl]oxy}phenyl)-2-(pyrimidin-5-yl)acetamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RHA
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with Z147647874 (Mpro-x2779) | | Descriptor: | 1-{4-[(thiophen-2-yl)methyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-05-15 | | Release date: | 2020-05-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen
To Be Published
|
|
5RJQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with NCL-00024665 | | Descriptor: | 2-(4-bromanyl-2-methoxy-phenyl)ethanoic acid, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RK5
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z57478994 | | Descriptor: | 5-(methoxymethyl)-1,3,4-thiadiazol-2-amine, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.243 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RKM
 
 | | PanDDA analysis group deposition -- Crystal Structure of PHIP in complex with Z2017168803 | | Descriptor: | (2S)-2-[(3-fluoropyridin-2-yl)(methyl)amino]propan-1-ol, PH-interacting protein | | Authors: | Grosjean, H, Aimon, A, Krojer, T, Talon, R, Douangamath, A, Koekemoer, L, Arrowsmith, C.H, Edwards, A, Bountra, C, von Delft, F, Biggin, P.C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5RL0
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-2 (Mpro-x3110) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ethyl N-[(2R)-2-[(4-tert-butylphenyl)(propanoyl)amino]-2-(pyridin-3-yl)acetyl]-beta-alaninate | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5REJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102241 | | Descriptor: | 1-{4-[(thiophen-2-yl)sulfonyl]piperazin-1-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2021-02-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5REZ
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with POB0129 | | Descriptor: | (1R,2S)-2-(thiophen-3-yl)cyclopentane-1-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5RFE
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z509756472 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(4-cyanophenyl)methyl]morpholine-4-carboxamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-03-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
6ZF9
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound SSR4 | | Descriptor: | 1,2-ETHANEDIOL, 3-ethyl-5-phenyl-[1,3]thiazolo[2,3-c][1,2,4]triazole, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound SSR4
To Be Published
|
|
2ZLE
 
 | | Cryo-EM structure of DegP12/OMP | | Descriptor: | Outer membrane protein C, Protease do | | Authors: | Schaefer, E, Saibil, H.R. | | Deposit date: | 2008-04-09 | | Release date: | 2008-06-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural basis for the regulated protease and chaperone function of DegP
Nature, 453, 2008
|
|
3MS5
 
 | | Crystal Structure of Human gamma-butyrobetaine,2-oxoglutarate dioxygenase 1 (BBOX1) | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-carboxyethyl)-1,1,1-trimethyldiazanium, Gamma-butyrobetaine dioxygenase, ... | | Authors: | Krojer, T, Kochan, G, McDonough, M.A, von Delft, F, Leung, I.K.H, Henry, L, Claridge, T.D.W, Pilka, E, Ugochukwu, E, Muniz, J, Filippakopoulos, P, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-04-29 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural and mechanistic studies on gamma-butyrobetaine hydroxylase.
Chem. Biol., 17, 2010
|
|
3O2G
 
 | | Crystal Structure of Human gamma-butyrobetaine,2-oxoglutarate dioxygenase 1 (BBOX1) | | Descriptor: | 1,2-ETHANEDIOL, 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, Gamma-butyrobetaine dioxygenase, ... | | Authors: | Krojer, T, Kochan, G, McDonough, M.A, von Delft, F, Leung, I.K.H, Henry, L, Claridge, T.D.W, Pilka, E, Ugochukwu, E, Muniz, J, Filippakopoulos, P, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K.L, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural and mechanistic studies on gamma-butyrobetaine hydroxylase.
Chem. Biol., 17, 2010
|
|
6ZEL
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F5 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-dimethyl-4-[(6-methylpyrimidin-4-yl)sulfanylmethyl]-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F5
To Be Published
|
|
6ZED
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F1 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-dimethyl-1~{H}-pyrazolo[3,4-c]pyridazin-3-amine, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F1
To Be Published
|
|
3QK3
 
 | | Crystal structure of human beta-crystallin B3 | | Descriptor: | 1,2-ETHANEDIOL, Beta-crystallin B3, SULFATE ION | | Authors: | Krojer, T, Gileadi, C, Cocking, R, Muniz, J, Pilka, E, Yue, W.W, Vollmar, M, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-01-31 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: |
|
|
4DO0
 
 | | Crystal Structure of human PHF8 in complex with Daminozide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DAMINOZIDE, ... | | Authors: | Krojer, T, Daniel, M, Ng, S.S, Walport, L.J, Chowdhury, R, Arrowsmith, C.H, Edwards, A, Bountra, C, Kawamura, A, Muller-Knapp, S, McDonough, M.A, von Delft, F, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: |
|
|
5JHJ
 
 | | M. Oryzae effector AVR-Pia mutant H3 | | Descriptor: | Antivirulence protein AVR-Pia | | Authors: | Padilla, A, deGuillen, K. | | Deposit date: | 2016-04-21 | | Release date: | 2017-03-29 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Recognition of the Magnaporthe oryzae Effector AVR-Pia by the Decoy Domain of the Rice NLR Immune Receptor RGA5.
Plant Cell, 29, 2017
|
|
5ZNE
 
 | |
5ZNG
 
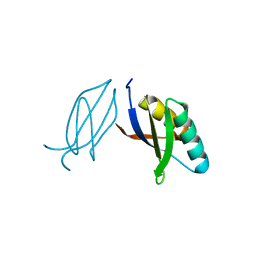 | | The crystal complex of immune receptor RGA5A_S of Pia from rice (Oryzae sativa) with rice blast (Magnaporthe oryzae) effector protein AVR1-CO39 | | Descriptor: | AVR1-CO39, NBS-LRR type protein | | Authors: | Guo, L.W, Zhang, Y.K, Liu, Q, Ma, M.Q, Liu, J.F, Peng, Y.L. | | Deposit date: | 2018-04-09 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Specific recognition of two MAX effectors by integrated HMA domains in plant immune receptors involves distinct binding surfaces
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
