7JZ4
 
 | |
7JZK
 
 | |
7JZ1
 
 | |
5OV7
 
 | | tubulin - rigosertib complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Menchon, G, Prota, A.E, Steinmetz, M, Jost, M. | | Deposit date: | 2017-08-28 | | Release date: | 2017-10-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent.
Mol. Cell, 68, 2017
|
|
7CQZ
 
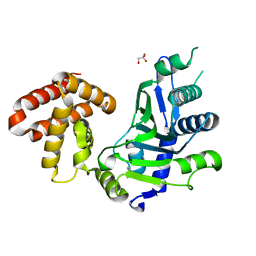 | | crystal structure of mouse FAM46C (TENT5C) | | Descriptor: | CHLORIDE ION, GLYCEROL, Terminal nucleotidyltransferase 5C | | Authors: | Hu, J.L, Zhang, H, Gao, S. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.34969759 Å) | | Cite: | Structural and functional characterization of multiple myeloma associated cytoplasmic poly(A) polymerase FAM46C.
Cancer Commun (Lond), 41, 2021
|
|
7EEA
 
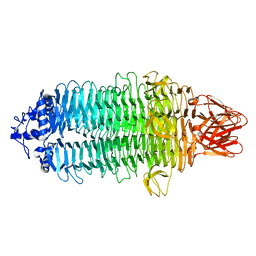 | |
7EEQ
 
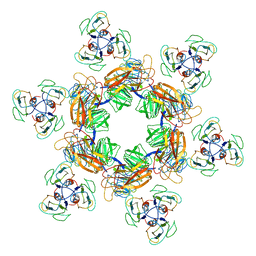 | | Cyanophage Pam1 tail machine | | Descriptor: | Needle head proteins, Tailspike head-binding domain | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEP
 
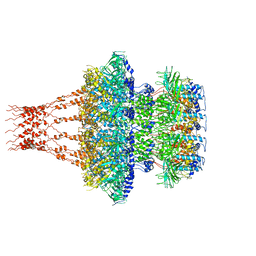 | | Cyanophage Pam1 portal-adaptor complex | | Descriptor: | Pam1 adaptor proteins, Pam1 portal proteins | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
7EEL
 
 | | Cyanophage Pam1 capsid asymmetric unit | | Descriptor: | Cement (decoration) proteins, Major capsid proteins | | Authors: | Zhang, J.T, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2021-03-19 | | Release date: | 2021-10-20 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure and assembly pattern of a freshwater short-tailed cyanophage Pam1.
Structure, 30, 2022
|
|
2W4D
 
 | | Acylphosphatase variant G91A from Pyrococcus horikoshii | | Descriptor: | ACYLPHOSPHATASE, PHOSPHATE ION, POTASSIUM ION | | Authors: | Lam, S.Y, Wong, K.B. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Rigidifying Salt-Bridge Favors the Activity of Thermophilic Enzyme at High Temperatures at the Expense of Low-Temperature Activity.
Plos Biol., 9, 2011
|
|
4XIM
 
 | |
4BSV
 
 | | Heterodimeric Fc Antibody Azymetric Variant 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, HETERODIMERIC FC ANTIBODY AZYMETRIC VARIANT 2, ... | | Authors: | Suits, M.D.L, Spreter, T, Cabrera, E.E, Dixit, S.B, Lario, P.I, Poon, D.K.Y, D'Angelo, I.E.P, Boulanger, M.J. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Improving Biophysical Properties of a Bispecific Antibody Scaffold to Aid Developability: Quality by Molecular Design.
Mabs, 5, 2013
|
|
8XIM
 
 | |
7F2E
 
 | | SARS-CoV-2 nucleocapsid protein C-terminal domain (dodecamer) | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Liu, C, Jiang, H. | | Deposit date: | 2021-06-10 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of the SARS-CoV-2 nucleocapsid protein C-terminal domain and development of nucleocapsid-targeting nanobodies.
Febs J., 289, 2022
|
|
7KCF
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-24512 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-hydroxyphenyl)-5-methyl-2-phenyl-3-(piperidin-1-yl)pyrazolo[1,5-a]pyrimidin-7(4H)-one, GLYCEROL, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KDA
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 34 | | Descriptor: | 2,3-diphenyl-5-[(1H-pyrazol-3-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KCE
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 2 | | Descriptor: | 5-methyl-2,3-diphenylpyrazolo[1,5-a]pyrimidin-7(4H)-one, CHLORIDE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KDB
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor compound 35 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-(4-hydroxyphenyl)-2,3-diphenyl-5-[(1H-pyrazol-3-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7KCC
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AG-270 | | Descriptor: | 1,2-ETHANEDIOL, 3-(cyclohex-1-en-1-yl)-6-(4-methoxyphenyl)-2-phenyl-5-[(pyridin-2-yl)amino]pyrazolo[1,5-a]pyrimidin-7(4H)-one, CHLORIDE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-10-05 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Discovery of AG-270, a First-in-Class Oral MAT2A Inhibitor for the Treatment of Tumors with Homozygous MTAP Deletion.
J.Med.Chem., 64, 2021
|
|
7CMO
 
 | | Crystal structure of human inorganic pyrophosphatase | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Hu, F, Huang, Z, Li, L. | | Deposit date: | 2020-07-28 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural and biochemical characterization of inorganic pyrophosphatase from Homo sapiens.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
4BSW
 
 | | Heterodimeric Fc Antibody Azymetric Variant 2 | | Descriptor: | 1,2-ETHANEDIOL, HETERODIMERIC FC ANTIBODY AZYMETRIC VARIANT 2, IODIDE ION, ... | | Authors: | Suits, M.D.L, Spreter, T, Cabrera, E.E, Dixit, S.B, Lario, P.I, Poon, D.K.Y, D'Angelo, I.E.P, Boulanger, M.J. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Improving Biophysical Properties of a Bispecific Antibody Scaffold to Aid Developability: Quality by Molecular Design.
Mabs, 5, 2013
|
|
7FJ2
 
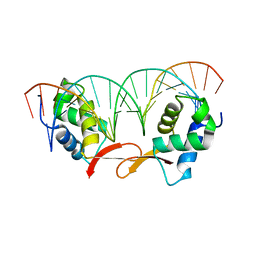 | | Structure of FOXM1 homodimer bound to a palindromic DNA site | | Descriptor: | DNA (5'-D(*AP*CP*CP*GP*TP*AP*AP*AP*CP*AP*TP*GP*TP*TP*TP*AP*CP*GP*GP*T)-3'), Forkhead box protein M1 | | Authors: | Dai, S.Y, Li, J, Zhang, H.J. | | Deposit date: | 2021-08-02 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.098 Å) | | Cite: | Mechanistic Insights into the Preference for Tandem Binding Sites in DNA Recognition by FOXM1.
J.Mol.Biol., 434, 2021
|
|
5UZZ
 
 | |
3L6T
 
 | |
5UZT
 
 | |
