2MLB
 
 | | NMR solution structure of a computational designed protein based on template of human erythrocytic ubiquitin | | Descriptor: | redesigned ubiquitin | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2MN4
 
 | | NMR solution structure of a computational designed protein based on structure template 1cy5 | | Descriptor: | Computational designed protein based on structure template 1cy5 | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-03-28 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2OBE
 
 | | Crystal Structure of Chimpanzee Adenovirus (Type 68/Simian 25) Major Coat Protein Hexon | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DIHYDROGENPHOSPHATE ION, Hexon protein | | Authors: | Xue, F, Rux, J.J, Burnett, R.M. | | Deposit date: | 2006-12-18 | | Release date: | 2007-07-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based identification of a major neutralizing site in an adenovirus hexon
J.Virol., 81, 2007
|
|
2QC3
 
 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
7YP1
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 10E4 (localized refinement) | | Descriptor: | 10E4 heavy chain, 10E4 light chain, EBV gH, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YP2
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAb 6H2 (localized refinement) | | Descriptor: | 6H2 heavy chain, 6H2 light chain, Envelope glycoprotein H | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7YOY
 
 | | Cryo-EM structure of EBV gHgL-gp42 in complex with mAbs 3E8 and 5E3 (localized refinement) | | Descriptor: | 3E8 heavy chain, 3E8 light chain, 5E3 heavy chain, ... | | Authors: | Liu, L, Sun, H, Jiang, Y, Hong, J, Zheng, Q, Li, S, Chen, Y, Xia, N. | | Deposit date: | 2022-08-02 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Non-overlapping epitopes on the gHgL-gp42 complex for the rational design of a triple-antibody cocktail against EBV infection.
Cell Rep Med, 4, 2023
|
|
7JG4
 
 | | Human GAR transformylase in complex with GAR substrate and AGF131 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-(5-{3-[(1S,7R,8R,9S)-4-amino-2-oxo-7lambda~4~-thia-3,5-diazatetracyclo[4.3.0.0~1,7~.0~7,9~]nona-3,5-dien-8-yl]propyl}thiophene-2-carbonyl)-L-glutamic acid, SODIUM ION, ... | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2020-07-18 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
7JG3
 
 | | Human GAR transformylase in complex with GAR substrate and AGF103 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]furan-2-carbonyl}-L-glutamic acid, SODIUM ION, ... | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2020-07-18 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
7JG0
 
 | | Human GAR transformylase in complex with GAR substrate and AGF102 inhibitor | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{5-[4-(2-amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]thiophene-2-carbonyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Wong-Roushar, J, Dann III, C.E. | | Deposit date: | 2020-07-18 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Discovery of 6-substituted thieno[2,3-d]pyrimidine analogs as dual inhibitors of glycinamide ribonucleotide formyltransferase and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase in de novo purine nucleotide biosynthesis in folate receptor expressing human tumors
Bioorg.Med.Chem., 37, 2021
|
|
6MO5
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50228 complex | | Descriptor: | MAGNESIUM ION, N-[(2S)-1-(hydroxyamino)-3-methyl-3-{[(oxetan-3-yl)methyl]sulfonyl}-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MOO
 
 | | Co-Crystal structure of P. aeruginosa LpxC-achn975 complex | | Descriptor: | N-[(2S)-3-azanyl-3-methyl-1-(oxidanylamino)-1-oxidanylidene-butan-2-yl]-4-[4-[(1R,2R)-2-(hydroxymethyl)cyclopropyl]buta -1,3-diynyl]benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MO4
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50067 complex | | Descriptor: | MAGNESIUM ION, N-[(2R)-1-(hydroxyamino)-3-methyl-3-(methylsulfonyl)-1-oxobutan-2-yl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, UDP-3-O-acyl-N-acetylglucosamine deacetylase | | Authors: | Stein, A.J, Assar, Z, Holt, M.C, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.844 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
6MOD
 
 | | Co-Crystal structure of P. aeruginosa LpxC-50432 complex | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-[(1S)-2-(hydroxyamino)-1-(3-methoxy-1,1-dioxo-1lambda~6~-thietan-3-yl)-2-oxoethyl]-4-(6-hydroxyhexa-1,3-diyn-1-yl)benzamide, ... | | Authors: | Stein, A.J, Holt, M.C, Assar, Z, Cohen, F, Andrews, L, Cirz, R. | | Deposit date: | 2018-10-04 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Optimization of LpxC Inhibitors for Antibacterial Activity and Cardiovascular Safety.
Chemmedchem, 14, 2019
|
|
7AMT
 
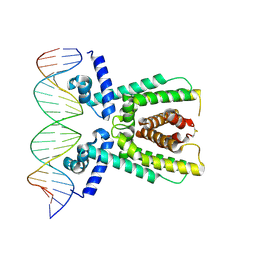 | | Structure of LuxR with DNA (activation) | | Descriptor: | DNA (5'-D(P*AP*TP*AP*AP*TP*GP*AP*CP*AP*TP*TP*AP*CP*TP*GP*TP*AP*TP*AP*TP*A)-3'), DNA (5'-D(P*TP*AP*TP*AP*TP*AP*CP*AP*GP*TP*AP*AP*TP*GP*TP*CP*AP*TP*TP*AP*T)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
7AMN
 
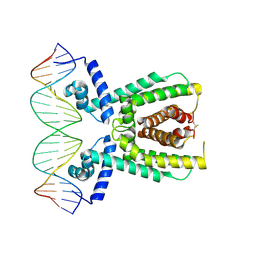 | | Structure of LuxR with DNA (repression) | | Descriptor: | DNA (5'-D(P*TP*AP*TP*TP*GP*AP*TP*AP*AP*AP*AP*TP*TP*AP*TP*CP*AP*AP*TP*AP*A)-3'), DNA (5'-D(P*TP*TP*AP*TP*TP*GP*AP*TP*AP*AP*TP*TP*TP*TP*AP*TP*CP*AP*AP*TP*A)-3'), HTH-type transcriptional regulator LuxR | | Authors: | Liu, B, Reverter, D. | | Deposit date: | 2020-10-09 | | Release date: | 2021-03-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Binding site profiles and N-terminal minor groove interactions of the master quorum-sensing regulator LuxR enable flexible control of gene activation and repression.
Nucleic Acids Res., 49, 2021
|
|
6MZN
 
 | |
3UGI
 
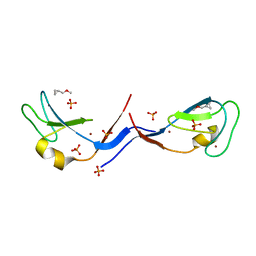 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | (methoxymethyl)cyclopropane, PHOSPHATE ION, Protein kinase C delta type, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | Structural and Functional Characterization of an Anesthetic Binding Site in the Second Cysteine-Rich Domain of Protein Kinase Cdelta
Biophys.J., 103, 2012
|
|
3UEJ
 
 | |
3UGL
 
 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | PHOSPHATE ION, Proteine kinase C delta type, ZINC ION, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.357 Å) | | Cite: | Structural and Functional Characterization of an Anesthetic Binding Site in the Second Cysteine-Rich Domain of Protein Kinase Cdelta
Biophys.J., 103, 2012
|
|
9JE9
 
 | | Crystal structure of a amidase that can hydrolase PU plastic | | Descriptor: | Amidase | | Authors: | Li, Z.S, Fang, S.T, Zheng, Z.R, Xia, W, Zhou, H.Y, Li, W.S, You, S, Han, X, Liu, W.D. | | Deposit date: | 2024-09-02 | | Release date: | 2025-07-09 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery and Engineering of a Urethanase for Enhanced Depolymerization of Polyurethane
Acs Catalysis, 15, 2025
|
|
6MZP
 
 | |
3UGD
 
 | | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Protein kinase C delta type, ... | | Authors: | Shanmugasundararaj, S, Stehle, T, Miller, K.W. | | Deposit date: | 2011-11-02 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and functional characterization of an anesthetic binding site in the second cysteine-rich domain of protein kinase C delta
Biophys.J., 103, 2012
|
|
3UFF
 
 | |
3UEY
 
 | |
