7DZJ
 
 | | Fabp protein before hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZK
 
 | | Fabp protein after hv | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7DZL
 
 | | A69C-M71L mutant of Fabp protein | | Descriptor: | Fatty acid-binding protein, liver, PALMITIC ACID | | Authors: | Li, H, Yu, L.-J, Liu, X, Shen, J.-R, Wang, J. | | Deposit date: | 2021-01-25 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Excited-state intermediates in a designer protein encoding a phototrigger caught by an X-ray free-electron laser.
Nat.Chem., 14, 2022
|
|
7RWH
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-41998 | | Descriptor: | 1,2-ETHANEDIOL, 8-(4-bromophenyl)-6-(4-methoxyphenyl)-2-[2,2,2-tris(fluoranyl)ethylamino]pyrido[4,3-d]pyrimidin-7-ol, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RWG
 
 | | "Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor AGI-43192 | | Descriptor: | (8R)-8-(4-chlorophenyl)-6-(2-methyl-2H-indazol-5-yl)-2-[(2,2,2-trifluoroethyl)amino]-5,8-dihydropyrido[4,3-d]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RW5
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 1 | | Descriptor: | (3'R)-N-(cyclopropylmethyl)-1'-[(2-fluorophenyl)methyl]-4-methyl-5H,7H-spiro[pyrano[4,3-d]pyrimidine-8,3'-pyrrolidin]-2-amine, S-ADENOSYLMETHIONINE, S-adenosylmethionine synthase isoform type-2 | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
7RW7
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Compound 9 | | Descriptor: | (3'R)-2-[(cyclopropylmethyl)amino]-6-(4-methoxyphenyl)-1'-[(1H-pyrazol-5-yl)methyl]-5,6-dihydro-7H-spiro[pyrido[4,3-d]pyrimidine-8,3'-pyrrolidin]-7-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Jin, L, Padyana, A.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Leveraging Structure-Based Drug Design to Identify Next-Generation MAT2A Inhibitors, Including Brain-Penetrant and Peripherally Efficacious Leads.
J.Med.Chem., 65, 2022
|
|
5E06
 
 | |
5E05
 
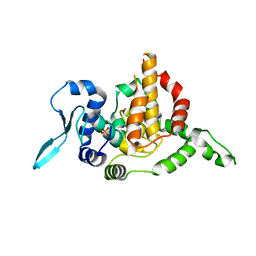 | |
2QXV
 
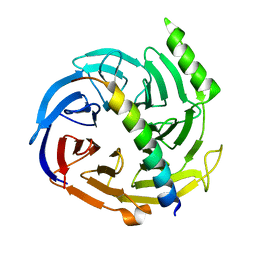 | | Structural basis of EZH2 recognition by EED | | Descriptor: | Embryonic ectoderm development, Enhancer of zeste homolog 2 | | Authors: | Han, Z. | | Deposit date: | 2007-08-13 | | Release date: | 2007-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural basis of EZH2 recognition by EED
Structure, 15, 2007
|
|
2OJ9
 
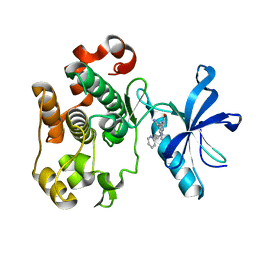 | | Structure of IGF-1R kinase domain complexed with a benzimidazole inhibitor | | Descriptor: | 3-[5-(1H-IMIDAZOL-1-YL)-7-METHYL-1H-BENZIMIDAZOL-2-YL]-4-[(PYRIDIN-2-YLMETHYL)AMINO]PYRIDIN-2(1H)-ONE, Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) | | Authors: | Sack, J.S, Jacobson, B.L. | | Deposit date: | 2007-01-12 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and initial SAR of 3-(1H-benzo[d]imidazol-2-yl)pyridin-2(1H)-ones as inhibitors of insulin-like growth factor 1-receptor (IGF-1R).
Bioorg.Med.Chem.Lett., 17, 2007
|
|
7SUG
 
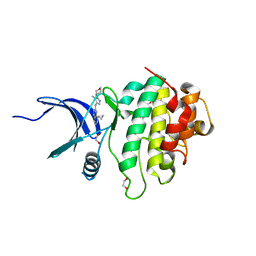 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 09 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{[5-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]amino}quinazolin-8-yl)cyclopropane-1-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SEO
 
 | | Crystal Structure of Caspase-3 with Peptide Inhibitor Ac-VDV(DAB)D-CHO | | Descriptor: | ACE-VAL-ASP-VAL-DAB-ASP, Caspase-3 subunit p12, Caspase-3 subunit p17 | | Authors: | Fuller, J.L, Finzel, B.C. | | Deposit date: | 2021-09-30 | | Release date: | 2022-01-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure-Based Design and Biological Evaluation of Novel Caspase-2 Inhibitors Based on the Peptide AcVDVAD-CHO and the Caspase-2-Mediated Tau Cleavage Sequence YKPVD314.
Acs Pharmacol Transl Sci, 5, 2022
|
|
7SUF
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 06 | | Descriptor: | 1,2-ETHANEDIOL, 8-cyclopropyl-N-[5-methyl-1-(oxan-4-yl)-1H-pyrazol-4-yl]quinazolin-2-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SUJ
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 24 | | Descriptor: | (3R,4R)-4-{4-[6-chloro-2-({1-[(1R)-2,2-difluorocyclopropyl]-5-methyl-1H-pyrazol-4-yl}amino)quinazolin-7-yl]piperidin-1-yl}-4-methyloxolan-3-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SUH
 
 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 15 | | Descriptor: | 1-[5-chloro-4-({6-chloro-7-[1-(oxetan-3-yl)piperidin-4-yl]quinazolin-2-yl}amino)-1H-pyrazol-1-yl]-2-methylpropan-2-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
7SUI
 
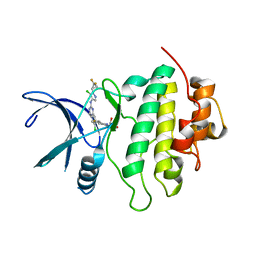 | | Structure of CHK1 10-pt. mutant complex with LRRK2 inhibitor 22 | | Descriptor: | (3R,4R)-4-{(3S,4S)-4-[6-chloro-2-({5-chloro-1-[(1R)-2,2-difluorocyclopropyl]-1H-pyrazol-4-yl}amino)quinazolin-7-yl]-3-fluoropiperidin-1-yl}oxolan-3-ol, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L. | | Deposit date: | 2021-11-17 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.119 Å) | | Cite: | Structure-Guided Discovery of Aminoquinazolines as Brain-Penetrant and Selective LRRK2 Inhibitors.
J.Med.Chem., 65, 2022
|
|
5E04
 
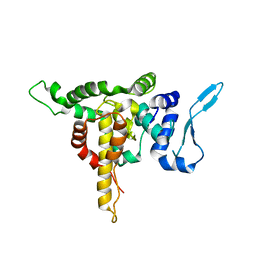 | | Crystal structure of Andes virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Wang, W.M, Lou, Z.Y. | | Deposit date: | 2015-09-28 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of the Core Region of Hantavirus Nucleocapsid Protein Reveals the Mechanism for Ribonucleoprotein Complex Formation
J.Virol., 90, 2015
|
|
9CTE
 
 | |
7U6R
 
 | |
7C01
 
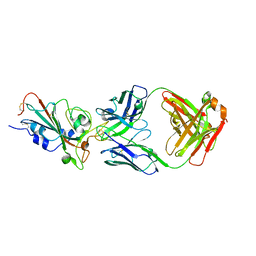 | | Molecular basis for a potent human neutralizing antibody targeting SARS-CoV-2 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CB6 heavy chain, CB6 light chain, ... | | Authors: | Shi, R, Qi, J, Wang, Q, Gao, F.G, Yan, J. | | Deposit date: | 2020-04-29 | | Release date: | 2020-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2.
Nature, 584, 2020
|
|
9CE4
 
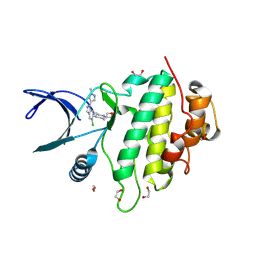 | | Structure of CHK1 10-pt. mutant complex with LRRK2 indazole inhibitor compound 6 | | Descriptor: | (1S,4r)-4-[(1P)-5-chloro-1-(1-methyl-1H-pyrazol-4-yl)-1H-indazol-6-yl]-1-(oxetan-3-yl)piperidin-1-ium, 1,2-ETHANEDIOL, Serine/threonine-protein kinase Chk1 | | Authors: | Palte, R.L, Zebisch, M, Henry, C, Barker, J.J. | | Deposit date: | 2024-06-26 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Discovery and Optimization of N-Heteroaryl Indazole LRRK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
7WPO
 
 | | Structure of NeoCOV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
7WPZ
 
 | | Structure of PDF-2180-COV RBD binding to Bat37 ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Cao, L, Wang, X, Tortorici, M.A, Veesler, D. | | Deposit date: | 2022-01-24 | | Release date: | 2022-11-30 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Close relatives of MERS-CoV in bats use ACE2 as their functional receptors.
Nature, 612, 2022
|
|
6KTW
 
 | | structure of EanB with hercynine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wu, L, Liu, P.H, Zhou, J.H. | | Deposit date: | 2019-08-29 | | Release date: | 2020-08-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.931 Å) | | Cite: | Single-Step Replacement of an Unreactive C-H Bond by a C-S Bond Using Polysulfide as the Direct Sulfur Source in the Anaerobic Ergothioneine Biosynthesis
Acs Catalysis, 10, 2020
|
|
