1VPE
 
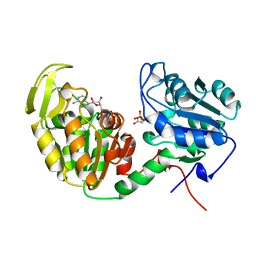 | | CRYSTALLOGRAPHIC ANALYSIS OF PHOSPHOGLYCERATE KINASE FROM THE HYPERTHERMOPHILIC BACTERIUM THERMOTOGA MARITIMA | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Auerbach, G, Huber, R, Graettinger, M, Zaiss, K, Schurig, H, Jaenicke, R, Jacob, U. | | Deposit date: | 1997-05-06 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Closed structure of phosphoglycerate kinase from Thermotoga maritima reveals the catalytic mechanism and determinants of thermal stability.
Structure, 5, 1997
|
|
1OB0
 
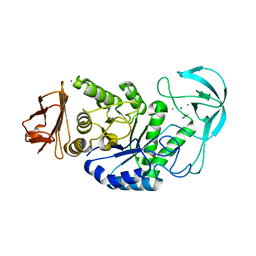 | | Kinetic stabilization of Bacillus licheniformis alpha-amylase through introduction of hydrophobic residues at the surface | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, SODIUM ION | | Authors: | Machius, M, Declerck, N, Huber, R, Wiegand, G. | | Deposit date: | 2003-01-21 | | Release date: | 2003-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Kinetic Stabilization of Bacillus Licheniformis Alpha-Amylase Through Introduction of Hydrophobic Residues at the Surface
J.Biol.Chem., 278, 2003
|
|
1STC
 
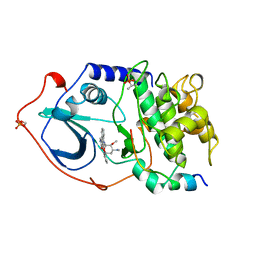 | | CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH STAUROSPORINE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR, STAUROSPORINE | | Authors: | Prade, L, Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1997-10-10 | | Release date: | 1998-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staurosporine-induced conformational changes of cAMP-dependent protein kinase catalytic subunit explain inhibitory potential.
Structure, 5, 1997
|
|
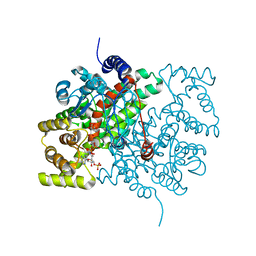
2CTS
 
 | |
3CSP
 
 | | Crystal structure of the DM2 mutant of myelin oligodendrocyte glycoprotein | | Descriptor: | Myelin-oligodendrocyte glycoprotein | | Authors: | Breithaupt, C, Schafer, B, Pellkofer, H, Huber, R, Linington, C, Jacob, U. | | Deposit date: | 2008-04-10 | | Release date: | 2008-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demyelinating myelin oligodendrocyte glycoprotein-specific autoantibody response is focused on one dominant conformational epitope region in rodents
J.Immunol., 181, 2008
|
|
1TSP
 
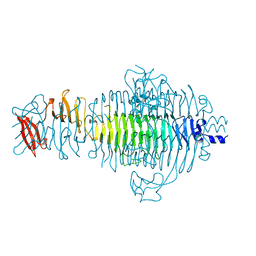 | | CRYSTAL STRUCTURE OF P22 TAILSPIKE PROTEIN: INTERDIGITATED SUBUNITS IN A THERMOSTABLE TRIMER | | Descriptor: | TAILSPIKE-PROTEIN | | Authors: | Steinbacher, S, Seckler, R, Miller, S, Steipe, B, Huber, R, Reinemer, P. | | Deposit date: | 1994-06-16 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer.
Science, 265, 1994
|
|
1TPO
 
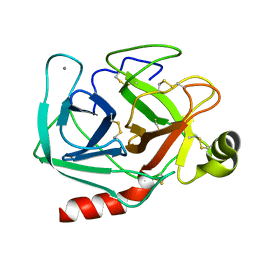 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | BETA-TRYPSIN, CALCIUM ION | | Authors: | Bode, W, Walter, J, Huber, R. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
1TPP
 
 | | THE GEOMETRY OF THE REACTIVE SITE AND OF THE PEPTIDE GROUPS IN TRYPSIN, TRYPSINOGEN AND ITS COMPLEXES WITH INHIBITORS | | Descriptor: | (2S)-3-(4-carbamimidoylphenyl)-2-hydroxypropanoic acid, BETA-TRYPSIN, CALCIUM ION, ... | | Authors: | Walter, J, Bode, W, Huber, R. | | Deposit date: | 1982-09-27 | | Release date: | 1983-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Geometry of the Reactive Site and of the Peptide Groups in Trypsin, Trypsinogen and its Complexes with Inhibitors
Acta Crystallogr.,Sect.B, 39, 1983
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
1M4Y
 
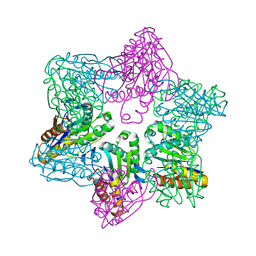 | | Crystal structure of HslV from Thermotoga maritima | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Song, H.K, Ramachandran, R, Bochtler, M.B, Hartmann, C, Azim, M.K, Huber, R. | | Deposit date: | 2002-07-05 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Isolation and characterization of the prokaryotic proteasome homolog HslVU (ClpQY) from Thermotoga maritima and the crystal structure of HslV.
BIOPHYS.CHEM., 100, 2003
|
|
1MVN
 
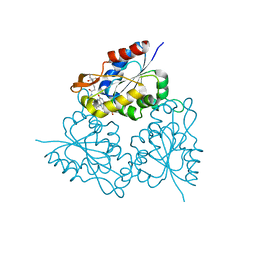 | | PPC decarboxylase mutant C175S complexed with pantothenoylaminoethenethiol | | Descriptor: | 2,4-DIHYDROXY-N-[2-(2-MERCAPTO-VINYLCARBAMOYL)-ETHYL]-3,3-DIMETHYL-BUTYRAMIDE, FLAVIN MONONUCLEOTIDE, PPC decarboxylase AtHAL3a | | Authors: | Steinbacher, S, Hernandez-Acosta, P, Bieseler, B, Blaesse, M, Huber, R, Culianez-Macia, F.A, Kupke, T. | | Deposit date: | 2002-09-26 | | Release date: | 2003-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of the Plant PPC Decarboxylase AtHAL3a Complexed with an Ene-thiol Reaction Intermediate
J.Mol.Biol., 327, 2003
|
|
1N6E
 
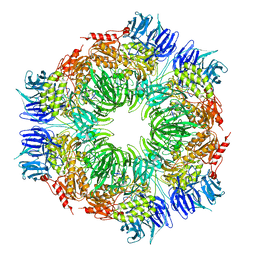 | | tricorn protease in complex with a tridecapeptide chloromethyl ketone derivative | | Descriptor: | DQTQKAAAELTFF, Tricorn protease | | Authors: | Kim, J.-S, Groll, M, Huber, R, Brandstetter, H. | | Deposit date: | 2002-11-10 | | Release date: | 2002-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Navigation Inside a Protease: Substrate Selection and Product Exit in the Tricorn Protease
from Thermoplasma acidophilum
J.Mol.Biol., 324, 2002
|
|
1WLH
 
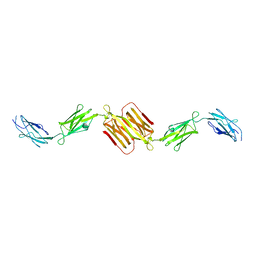 | | Molecular structure of the rod domain of Dictyostelium filamin | | Descriptor: | Gelation factor | | Authors: | Popowicz, G.M, Mueller, R, Noegel, A.A, Schleicher, M, Huber, R, Holak, T.A. | | Deposit date: | 2004-06-27 | | Release date: | 2004-10-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular structure of the rod domain of dictyostelium filamin
J.Mol.Biol., 342, 2004
|
|
1N6F
 
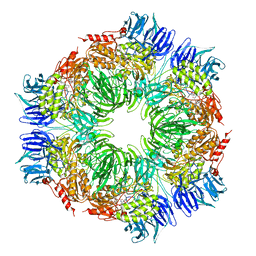 | | tricorn protease in complex with Z-Phe-diketo-Arg-Glu-Phe | | Descriptor: | 4-[2-(3-BENZYLOXYCARBONYLAMINO-4-CYCLOHEXYL-1-HYDROXY-2-OXO-BUTYLAMINO)-5-GUANIDINO-PENTANOYLAMINO]-4-(1-CARBOXY-2-CYCLOHEXYL-ETHYLCARBAMOYL)-BUTYRIC ACID, tricorn protease | | Authors: | Kim, J.-S, Groll, M, Huber, R, Brandstetter, H. | | Deposit date: | 2002-11-10 | | Release date: | 2002-12-30 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Navigation Inside a Protease: Substrate Selection and Product Exit in the Tricorn Protease
from Thermoplasma acidophilum
J.Mol.Biol., 324, 2002
|
|
1N6D
 
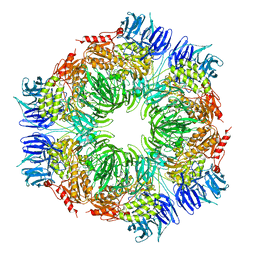 | | Tricorn protease in complex with tetrapeptide chloromethyl ketone derivative | | Descriptor: | DECANE, RVRK, Tricorn protease | | Authors: | Kim, J.-S, Groll, M, Huber, R, Brandstetter, H. | | Deposit date: | 2002-11-10 | | Release date: | 2002-12-30 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Navigation Inside a Protease: Substrate Selection and Product Exit in the Tricorn Protease
from Thermoplasma acidophilum
J.Mol.Biol., 324, 2002
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
2FB4
 
 | | DIR PRIMAERSTRUKTUR DES KRISTALLISIERBAREN MONOKLONALEN IMMUNOGLOBULINS IGG1 KOL. II. AMINOSAEURESEQUENZ DER L-KETTE, LAMBDA-TYP, SUBGRUPPE I (GERMAN) | | Descriptor: | IGG1-LAMBDA KOL FAB (HEAVY CHAIN), IGG1-LAMBDA KOL FAB (LIGHT CHAIN) | | Authors: | Marquart, M, Huber, R. | | Deposit date: | 1989-04-18 | | Release date: | 1989-07-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The primary structure of crystallizable monoclonal immunoglobulin IgG1 Kol. II. Amino acid sequence of the L-chain, gamma-type, subgroup I
Biol.Chem.Hoppe-Seyler, 370, 1989
|
|
1VJW
 
 | | STRUCTURE OF OXIDOREDUCTASE (NADP+(A),FERREDOXIN(A)) | | Descriptor: | FERREDOXIN(A), IRON/SULFUR CLUSTER | | Authors: | Macedo-Ribeiro, S, Darimont, B, Sterner, R, Huber, R. | | Deposit date: | 1996-10-09 | | Release date: | 1996-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small structural changes account for the high thermostability of 1[4Fe-4S] ferredoxin from the hyperthermophilic bacterium Thermotoga maritima.
Structure, 4, 1996
|
|
1MVL
 
 | | PPC decarboxylase mutant C175S | | Descriptor: | FLAVIN MONONUCLEOTIDE, PPC decarboxylase AtHAL3a | | Authors: | Steinbacher, S, Hernandez-Acosta, P, Bieseler, B, Blaesse, M, Huber, R, Culianez-Macia, F.A, Kupke, T. | | Deposit date: | 2002-09-26 | | Release date: | 2003-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Plant PPC Decarboxylase AtHAL3a Complexed with an Ene-thiol Reaction Intermediate
J.Mol.Biol., 327, 2003
|
|
1JAO
 
 | | COMPLEX OF 3-MERCAPTO-2-BENZYLPROPANOYL-ALA-GLY-NH2 WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), N-[(2S)-2-benzyl-3-sulfanylpropanoyl]-L-alanylglycinamide, ... | | Authors: | Grams, F, Reinemer, P, Powers, J.C, Kleine, T, Piper, M, Tschesche, H, Huber, R, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design.
Eur.J.Biochem., 228, 1995
|
|
1JAN
 
 | | COMPLEX OF PRO-LEU-GLY-HYDROXYLAMINE WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (PHE79 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (PHE79 FORM), PRO-LEU-GLY-HYDROXYLAMINE INHIBITOR, ... | | Authors: | Reinemer, P, Grams, F, Huber, R, Kleine, T, Schnierer, S, Pieper, M, Tschesche, H, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural implications for the role of the N terminus in the 'superactivation' of collagenases. A crystallographic study.
FEBS Lett., 338, 1994
|
|
1JAP
 
 | | COMPLEX OF PRO-LEU-GLY-HYDROXYLAMINE WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), PRO-LEU-GLY-HYDROXYLAMINE, ... | | Authors: | Bode, W, Reinemer, P, Huber, R, Kleine, T, Schnierer, S, Tschesche, H. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The X-ray crystal structure of the catalytic domain of human neutrophil collagenase inhibited by a substrate analogue reveals the essentials for catalysis and specificity.
EMBO J., 13, 1994
|
|
2QZX
 
 | | Secreted aspartic proteinase (Sap) 5 from Candida albicans | | Descriptor: | Candidapepsin-5, Pepstatin | | Authors: | Lee, J.H, Ruge, E, Borelli, C, Maskos, K, Huber, R. | | Deposit date: | 2007-08-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structures of Sap1 and Sap5: Structural comparison of the secreted aspartic proteinases from Candida albicans.
Proteins, 72, 2008
|
|
1JC9
 
 | | TACHYLECTIN 5A FROM TACHYPLEUS TRIDENTATUS (JAPANESE HORSESHOE CRAB) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, techylectin-5A | | Authors: | Kairies, N, Beisel, H.-G, Fuentes-Prior, P, Tsuda, R, Muta, T, Iwanaga, S, Bode, W, Huber, R, Kawabata, S. | | Deposit date: | 2001-06-08 | | Release date: | 2001-11-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The 2.0-A crystal structure of tachylectin 5A provides evidence for the common origin of the innate immunity and the blood coagulation systems.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1JAQ
 
 | | COMPLEX OF 1-HYDROXYLAMINE-2-ISOBUTYLMALONYL-ALA-GLY-NH2 WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), N-[(2R)-2-(hydroxycarbamoyl)-4-methylpentanoyl]-L-alanylglycinamide, ... | | Authors: | Grams, F, Reinemer, P, Powers, J.C, Kleine, T, Piper, M, Tschesche, H, Huber, R, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design.
Eur.J.Biochem., 228, 1995
|
|
