6E1Z
 
 | |
6E23
 
 | |
6E22
 
 | |
8H77
 
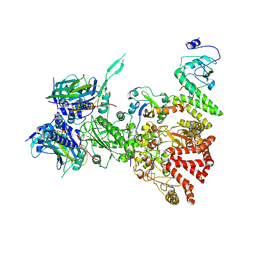 | | Hsp90-AhR-p23-XAP2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
7JQ3
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI6 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ0
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI3 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ1
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI4 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ2
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI5 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-alaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JPY
 
 | |
7JQ4
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI7 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-threonyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JPZ
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI1 | | Descriptor: | (phenylmethyl) N-[(2S)-1-oxidanylidene-1-[[(2S)-1-oxidanyl-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-3-phenyl-propan-2-yl]carbamate, 3C-like proteinase | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7JQ5
 
 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI8 | | Descriptor: | 3C-like proteinase, N-[(BENZYLOXY)CARBONYL]-O-(TERT-BUTYL)-L-THREONYL-3-CYCLOHEXYL-N-[(1S)-2-HYDROXY-1-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}ETHYL]-L-ALANINAMIDE | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
4JGV
 
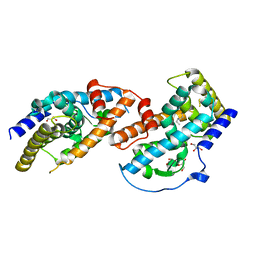 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | Deposit date: | 2013-03-04 | | Release date: | 2013-12-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
8FXS
 
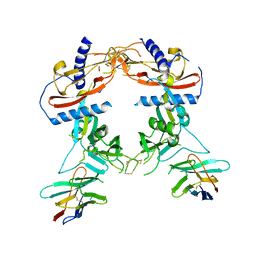 | | Crystal structure of human pro-TGF-beta2 in complex with Nb9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody clone 9, Transforming growth factor beta-2 proprotein | | Authors: | Le, V.Q, Springer, T.A. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-21 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A specialized integrin-binding motif enables proTGF-beta 2 activation by integrin alpha V beta 6 but not alpha V beta 8.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FXV
 
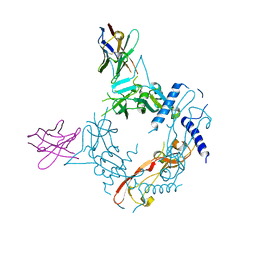 | | Crystal structure of human proTGF-beta2 in complex with Nb18 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody clone 18, Transforming growth factor beta-2 proprotein | | Authors: | Le, V.Q, Springer, T.A. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A specialized integrin-binding motif enables proTGF-beta 2 activation by integrin alpha V beta 6 but not alpha V beta 8.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8V8J
 
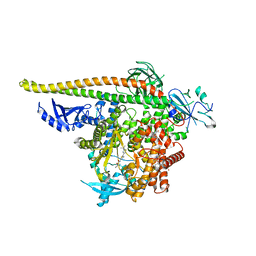 | | PI3Ka H1047R co-crystal structure with inhibitors in two cryptic pockets (compounds 4 and 5). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, 2-({(1R)-1-[2-(4,4-dimethylpiperidin-1-yl)-3,6-dimethyl-4-oxo-4H-1-benzopyran-8-yl]ethyl}amino)benzoic acid, N-{(3S)-3-(2-methylphenyl)-6-[(oxetan-3-yl)amino]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-1-benzothiophene-3-carboxamide, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8V8H
 
 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket near H1047R (compound 4). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, 2-({(1R)-1-[2-(4,4-dimethylpiperidin-1-yl)-3,6-dimethyl-4-oxo-4H-1-benzopyran-8-yl]ethyl}amino)benzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.58 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8V8V
 
 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket near H1047R (compound 7). | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, 2-[[(1~{R})-1-(7-methyl-4-oxidanylidene-2-piperidin-1-yl-3~{H}-pyrido[1,2-a]pyrimidin-9-yl)ethyl]amino]benzoic acid, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8V8I
 
 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket (compound 5). | | Descriptor: | (2S)-N~1~-[(4P)-2-tert-butyl-4'-methyl[4,5'-bi-1,3-thiazol]-2'-yl]pyrrolidine-1,2-dicarboxamide, CHLORIDE ION, N-{(3S)-3-(2-methylphenyl)-6-[(oxetan-3-yl)amino]-1-oxo-2,3-dihydro-1H-isoindol-4-yl}-1-benzothiophene-3-carboxamide, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
8V8U
 
 | | PI3Ka H1047R co-crystal structure with inhibitor in cryptic pocket near H1047R (compound 12). | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, (3S)-9-[(1R)-1-(2-carboxyanilino)ethyl]-3-cyano-7-methyl-4-oxo-2-(piperidin-1-yl)-3,4-dihydropyrido[1,2-a]pyrimidin-5-ium, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Gunn, R.J, Lawson, J.D. | | Deposit date: | 2023-12-06 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Discovery of Pyridopyrimidinones that Selectively Inhibit the H1047R PI3K alpha Mutant Protein.
J.Med.Chem., 67, 2024
|
|
5V8V
 
 | | Crystal Structure of Human Renin in Complex with a biphenylpipderidinylcarbinol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin, methyl [(4S)-4-(3'-ethyl-6-fluoro[1,1'-biphenyl]-2-yl)-4-hydroxy-4-{(3R)-1-[4-(methylamino)butanoyl]piperidin-3-yl}butyl]carbamate | | Authors: | Concha, N, Zhao, B. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of renin inhibitors containing a simple aspartate binding moiety that imparts reduced P450 inhibition.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3N75
 
 | | X-ray Crystal Structure of the Escherichia coli Inducible Lysine Decarboxylase LdcI | | Descriptor: | GLYCEROL, GUANOSINE-5',3'-TETRAPHOSPHATE, HEXAETHYLENE GLYCOL, ... | | Authors: | Kanjee, U, Alexopoulos, E, Pai, E.F, Houry, W.A. | | Deposit date: | 2010-05-26 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase.
Embo J., 30, 2011
|
|
3NBX
 
 | |
5VPM
 
 | |
5VRP
 
 | |
