8XFH
 
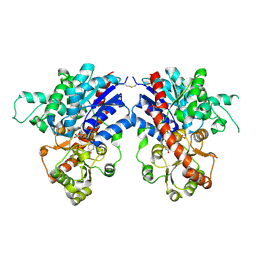 | |
7DPM
 
 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with MW06 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Wang, J, Jiao, S, Wang, R, Zhang, J, Zhang, M, Wang, M. | | Deposit date: | 2020-12-20 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.304 Å) | | Cite: | Characterization of MW06, a human monoclonal antibody with cross-neutralization activity against both SARS-CoV-2 and SARS-CoV.
Mabs, 13, 2021
|
|
4FZ4
 
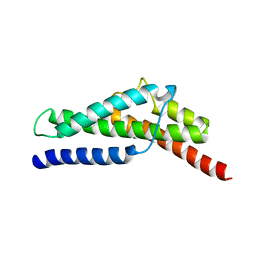 | | Crystal structure of HP0197-18kd | | Descriptor: | CHLORIDE ION, NITRATE ION, Uncharacterized protein conserved in bacteria | | Authors: | Yuan, Z, Yan, X. | | Deposit date: | 2012-07-06 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Molecular mechanism by which surface antigen HP0197 mediates host cell attachment in the pathogenic bacteria Streptococcus suis
J.Biol.Chem., 288, 2013
|
|
4FZQ
 
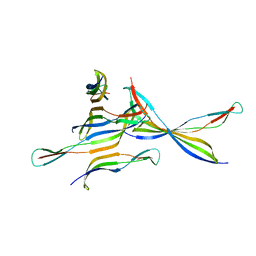 | | Crystal structure of HP0197-G5 | | Descriptor: | Uncharacterized protein conserved in bacteria | | Authors: | Yuan, Z, Yan, X. | | Deposit date: | 2012-07-07 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism by which surface antigen HP0197 mediates host cell attachment in the pathogenic bacteria Streptococcus suis
J.Biol.Chem., 288, 2013
|
|
4FIN
 
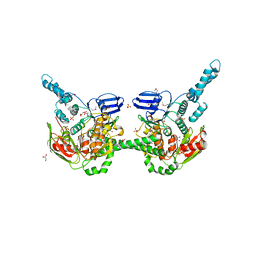 | | Crystal Structure of EttA (formerly YjjK) - an E. coli ABC-type ATPase | | Descriptor: | CITRIC ACID, EttA (YjjK) ABCF family protein, GLYCEROL, ... | | Authors: | Smith, P, Yuan, Y, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-09 | | Release date: | 2013-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The ABC-F protein EttA gates ribosome entry into the translation elongation cycle.
Nat.Struct.Mol.Biol., 21, 2014
|
|
8HS3
 
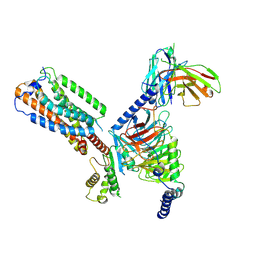 | | Gi bound orphan GPR20 in ligand-free state | | Descriptor: | Ggama, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HSC
 
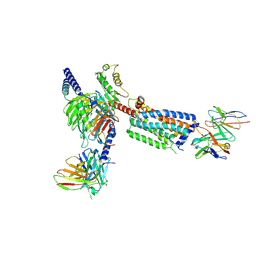 | | Gi bound Orphan GPR20 complex with Fab046 in ligand-free state | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-19 | | Release date: | 2023-03-08 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8HS2
 
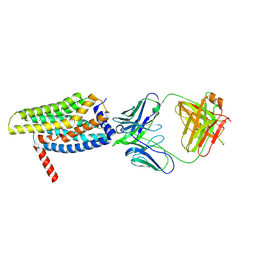 | | Orphan GPR20 in complex with Fab046 | | Descriptor: | Light chain of Fab046, Soluble cytochrome b562,G-protein coupled receptor 20, heavy chain of Fab046 | | Authors: | Lin, X, Jiang, S, Xu, F. | | Deposit date: | 2022-12-16 | | Release date: | 2023-03-08 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | The activation mechanism and antibody binding mode for orphan GPR20.
Cell Discov, 9, 2023
|
|
8I2J
 
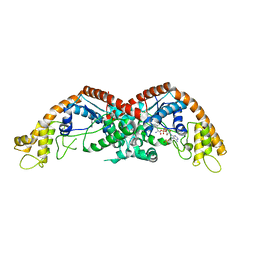 | |
8I2M
 
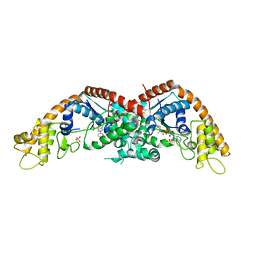 | |
8I1Y
 
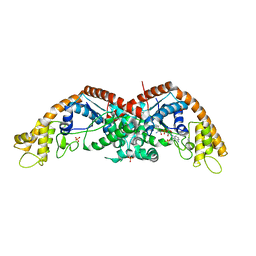 | | The structure of E. coli TrpRS bound with a chemical fragment | | Descriptor: | 5-ethanoylthiophene-2-carbonitrile, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
8I27
 
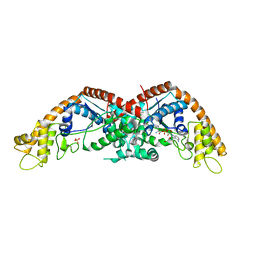 | |
8I2A
 
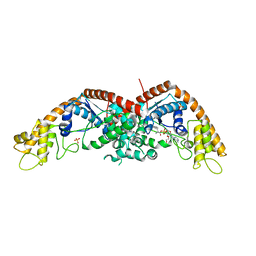 | |
8I4I
 
 | |
8I1W
 
 | |
8I2L
 
 | |
8I2C
 
 | |
8I1Z
 
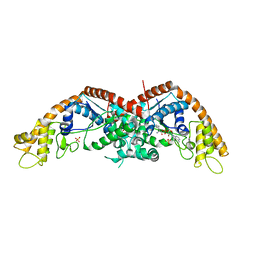 | | E. coli tryptophanyl-tRNA synthetase bound with a chemical fragment | | Descriptor: | 1-(2,3-dihydro-1-benzofuran-5-yl)ethanone, SULFATE ION, TRYPTOPHANYL-5'AMP, ... | | Authors: | Xiang, M, Zhou, H. | | Deposit date: | 2023-01-13 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An asymmetric structure of bacterial TrpRS supports the half-of-the-sites catalytic mechanism and facilitates antimicrobial screening.
Nucleic Acids Res., 51, 2023
|
|
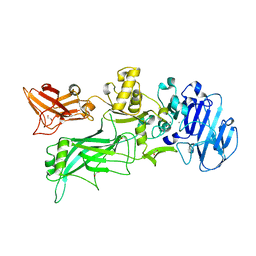
4H2A
 
 | |
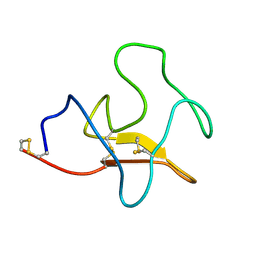
1KIV
 
 | |
7TZL
 
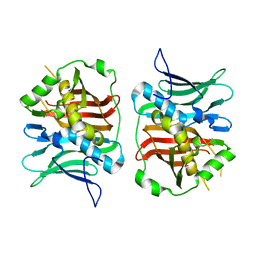 | | The DH dehydratase domain of AlnB | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Swain, K, Blackson, W, Wang, B, Zhao, H, Nannenga, B.L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The programming of alpha,beta-polyene biosynthesis by a bacterial iterative type I polyketide synthase
To Be Published
|
|
7VA8
 
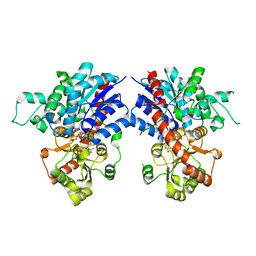 | | Crystal structure of MiCGT | | Descriptor: | UDP-glycosyltransferase 13, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Zhong, L, Zhang, Z.M. | | Deposit date: | 2021-08-27 | | Release date: | 2022-07-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85003233 Å) | | Cite: | Directed Evolution of a Plant Glycosyltransferase for Chemo- and Regioselective Glycosylation of Pharmaceutically Significant Flavonoids
Acs Catalysis, 11, 2021
|
|
7VAA
 
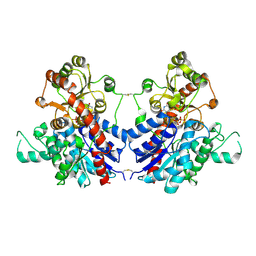 | |
7YSE
 
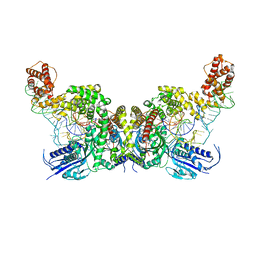 | | Crystal structure of E. coli heterotetrameric GlyRS in complex with tRNA | | Descriptor: | Glycine--tRNA ligase alpha subunit, Glycine--tRNA ligase beta subunit, MAGNESIUM ION, ... | | Authors: | Han, L, Ju, Y, Zhou, H. | | Deposit date: | 2022-08-12 | | Release date: | 2023-02-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.907 Å) | | Cite: | The binding mode of orphan glycyl-tRNA synthetase with tRNA supports the synthetase classification and reveals large domain movements.
Sci Adv, 9, 2023
|
|
6JBT
 
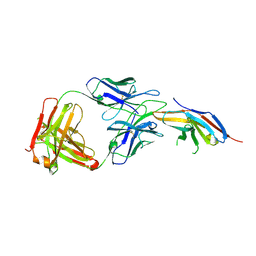 | | Complex structure of toripalimab-Fab and PD-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain, ... | | Authors: | Guo, L, Tan, S, Chai, Y, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2019-01-26 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Glycosylation-independent binding of monoclonal antibody toripalimab to FG loop of PD-1 for tumor immune checkpoint therapy.
Mabs, 11, 2019
|
|
