1FTC
 
 | | Y13C MUTANT OF AZOTOBACTER VINELANDII FDI | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Kemper, M.A, Lloyd, S.J, Prasad, G.S, Stout, C.D, Fawcett, S, Armstrong, F.A, Burgess, B.K. | | Deposit date: | 1997-01-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Y13C Azotobacter vinelandii ferredoxin I. A designed [Fe-S] ligand motif contains a cysteine persulfide.
J.Biol.Chem., 272, 1997
|
|
1FGH
 
 | | COMPLEX WITH 4-HYDROXY-TRANS-ACONITATE | | Descriptor: | 4-HYDROXY-ACONITATE ION, ACONITASE, IRON/SULFUR CLUSTER | | Authors: | Lauble, H, Kennedy, M.C, Emptage, M.H, Beinert, H, Stout, C.D. | | Deposit date: | 1996-09-14 | | Release date: | 1996-12-23 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The reaction of fluorocitrate with aconitase and the crystal structure of the enzyme-inhibitor complex.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1GAK
 
 | |
1G1X
 
 | | STRUCTURE OF RIBOSOMAL PROTEINS S15, S6, S18, AND 16S RIBOSOMAL RNA | | Descriptor: | 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S15, 30S RIBOSOMAL PROTEIN S18, ... | | Authors: | Agalarov, S.C, Prasad, G.S, Funke, P.M, Stout, C.D, Williamson, J.R. | | Deposit date: | 2000-10-13 | | Release date: | 2000-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain.
Science, 288, 2000
|
|
1EGK
 
 | | CRYSTAL STRUCTURE OF A NUCLEIC ACID FOUR-WAY JUNCTION | | Descriptor: | 10-23 DNA ENZYME, MAGNESIUM ION, RNA (5'-R(*AP*GP*GP*AP*GP*AP*GP*AP*GP*AP*UP*GP*GP*GP*UP*GP*CP*GP*AP*G)-3') | | Authors: | Nowakowski, J, Shim, P.J, Stout, C.D, Joyce, G.F. | | Deposit date: | 2000-02-15 | | Release date: | 2000-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Alternative conformations of a nucleic acid four-way junction.
J.Mol.Biol., 300, 2000
|
|
1F5C
 
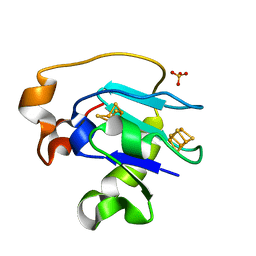 | | CRYSTAL STRUCTURE OF F25H FERREDOXIN 1 MUTANT FROM AZOTOBACTER VINELANDII AT 1.75 ANGSTROM RESOLUTION | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER, ... | | Authors: | Chen, K, Bonagura, C.A, Tilley, G.J, Jung, Y.S, Armstrong, F.A, Stout, C.D, Burgess, B.K. | | Deposit date: | 2000-06-13 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential.
Nat.Struct.Biol., 9, 2002
|
|
1F5B
 
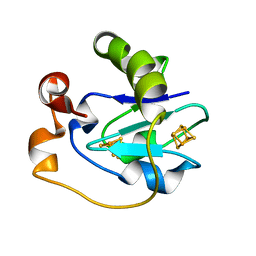 | | CRYSTAL STRUCTURE OF F2H FERREDOXIN 1 MUTANT FROM AZOTOBACTER VINELANDII AT 1.75 ANGSTROM RESOLUTION | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER | | Authors: | Chen, K, Bonagura, C.A, Tilley, G.J, Jung, Y.S, Armstrong, F.A, Stout, C.D, Burgess, B.K. | | Deposit date: | 2000-06-13 | | Release date: | 2000-06-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal structures of ferredoxin variants exhibiting large changes in [Fe-S] reduction potential.
Nat.Struct.Biol., 9, 2002
|
|
3KFR
 
 | |
3KFP
 
 | |
3KF0
 
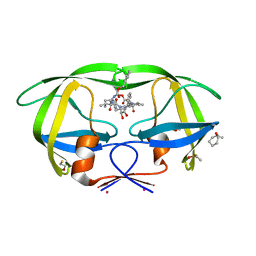 | | HIV Protease with fragment 4D9 bound | | Descriptor: | (1S,2S)-2-methylcyclohexanol, BETA-MERCAPTOETHANOL, DIMETHYL SULFOXIDE, ... | | Authors: | Stout, C.D, Perryman, A.L. | | Deposit date: | 2009-10-27 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based screen against HIV protease.
Chem.Biol.Drug Des., 75, 2010
|
|
3KFS
 
 | |
3KFN
 
 | |
3EH4
 
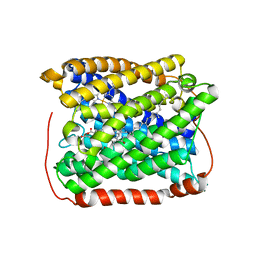 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
3EH3
 
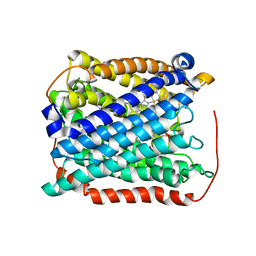 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
3EH5
 
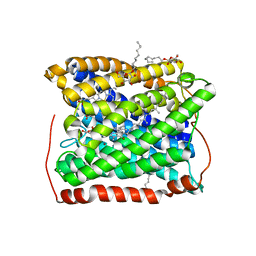 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
5MR3
 
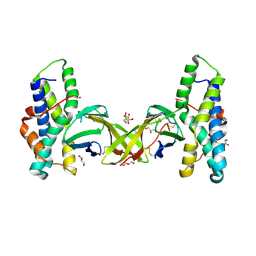 | | Crystal structure of red abalone egg VERL repeat 2 with linker in complex with sperm lysin at 1.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Egg-lysin, ... | | Authors: | Nishimura, K, Raj, I, Sadat Al-Hosseini, H, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-12-21 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
1TRA
 
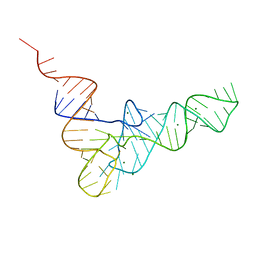 | | RESTRAINED REFINEMENT OF THE MONOCLINIC FORM OF YEAST PHENYLALANINE TRANSFER RNA. TEMPERATURE FACTORS AND DYNAMICS, COORDINATED WATERS, AND BASE-PAIR PROPELLER TWIST ANGLES | | Descriptor: | MAGNESIUM ION, TRNAPHE | | Authors: | Westhof, E, Sundaralingam, M. | | Deposit date: | 1986-05-16 | | Release date: | 1986-07-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Restrained refinement of the monoclinic form of yeast phenylalanine transfer RNA. Temperature factors and dynamics, coordinated waters, and base-pair propeller twist angles.
Biochemistry, 25, 1986
|
|
5IIB
 
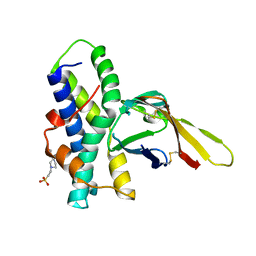 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.64 A resolution (crystal form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Egg-lysin, ... | | Authors: | Raj, I, Sadat Al-Hosseini, H, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5II7
 
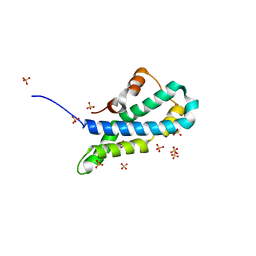 | | In-house sulfur-SAD structure of orthorhombic red abalone lysin at 1.66 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Egg-lysin, SULFATE ION | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5II9
 
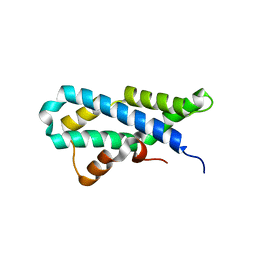 | |
5IIA
 
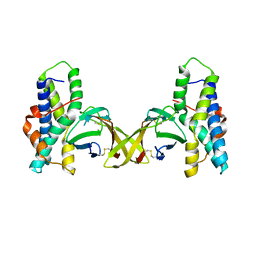 | | Crystal structure of red abalone egg VERL repeat 3 in complex with sperm lysin at 1.7 A resolution (crystal form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Egg-lysin, Vitelline envelope sperm lysin receptor | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
5II8
 
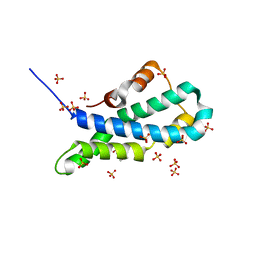 | | Orthorhombic crystal structure of red abalone lysin at 0.99 A resolution | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Egg-lysin, SULFATE ION | | Authors: | Sadat Al-Hosseini, H, Raj, I, Nishimura, K, De Sanctis, D, Jovine, L. | | Deposit date: | 2016-03-01 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Structural Basis of Egg Coat-Sperm Recognition at Fertilization.
Cell, 169, 2017
|
|
4TVH
 
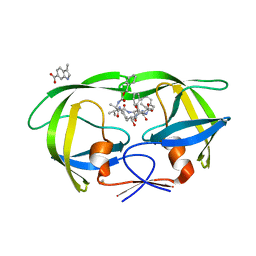 | | HIV Protease (PR) dimer in closed form with TL-3 in active site and fragment AK-2097 in the outside/top of flap | | Descriptor: | 3-(morpholin-4-ylmethyl)-1H-indole-6-carboxylic acid, BETA-MERCAPTOETHANOL, Protease, ... | | Authors: | Tiefenbrunn, T, Kislukhin, A, Finn, M.G, Stout, C.D. | | Deposit date: | 2014-06-26 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.195 Å) | | Cite: | Distinguishing Binders from False Positives by Free Energy Calculations: Fragment Screening Against the Flap Site of HIV Protease.
J.Phys.Chem.B, 119, 2015
|
|
3IBD
 
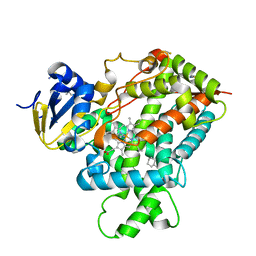 | | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Gay, S.C, Sun, L, Talakad, J.C, Shah, M.B, Stout, D.C, Halpert, J.R. | | Deposit date: | 2009-07-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cytochrome P450 2B6 genetic variant in complex with the inhibitor 4-(4-chlorophenyl)imidazole at 2.0-A resolution.
Mol.Pharmacol., 77, 2010
|
|
4TVG
 
 | | HIV Protease (PR) dimer in closed form with pepstatin in active site and fragment AK-2097 in the outside/top of flap | | Descriptor: | 3-(morpholin-4-ylmethyl)-1H-indole-6-carboxylic acid, GLYCEROL, HIV Protease, ... | | Authors: | Tiefenbrunn, T, Kislukhin, A, Finn, M.G, Stout, C.D. | | Deposit date: | 2014-06-26 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Distinguishing Binders from False Positives by Free Energy Calculations: Fragment Screening Against the Flap Site of HIV Protease.
J.Phys.Chem.B, 119, 2015
|
|
