8TKN
 
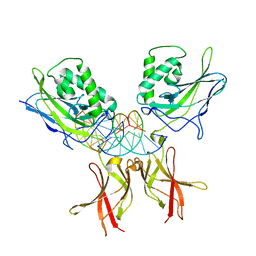 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with 10-mer kappaB DNA from human Neutrophil Gelatinase-associated Lipocalin (NGAL) promoter | | Descriptor: | DNA A, DNA B, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Zhu, N, Mealka, M, Mitchel, S, Rogers, W.E, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
8TKM
 
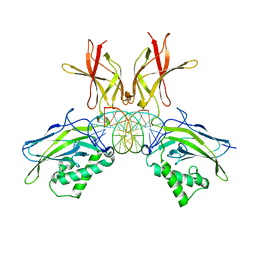 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with 17-mer kappaB DNA from human interleukin-6 (IL-6) promoter | | Descriptor: | 17-mer kappaB DNA, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Zhu, N, Mealka, M, Mitchel, S, Rogers, W.E, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
5JK5
 
 | |
5JK6
 
 | |
5JK8
 
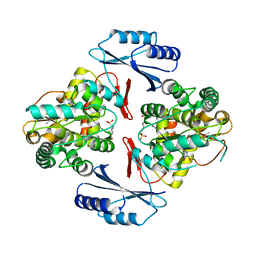 | |
6JUZ
 
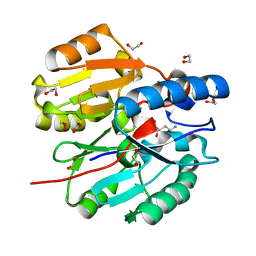 | | Crystal Structure of N-terminal domain of ArgZ(N71S) covalently bond to a reaction intermediate | | Descriptor: | 1,2-ETHANEDIOL, ARGININE, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhuang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
4HAC
 
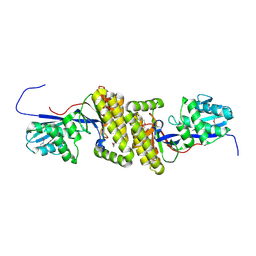 | |
4HU0
 
 | |
6JV0
 
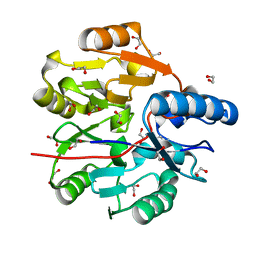 | | Crystal Structure of N-terminal domain of ArgZ, bound to Product, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | 1,2-ETHANEDIOL, L-ornithine, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6JUY
 
 | | Crystal Structure of ArgZ, apo structure, an Arginine Dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
6JV1
 
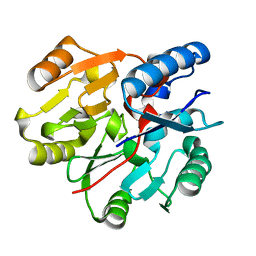 | | Crystal Structure of N-terminal domain of ArgZ, C264S mutant, bound to Substrate, an arginine dihydrolase from the Ornithine-Ammonia Cycle in Cyanobacteria | | Descriptor: | ARGININE, Sll1336 protein | | Authors: | Zhuang, N, Li, L, Wu, X, Zhang, Y. | | Deposit date: | 2019-04-15 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structures and biochemical analyses of the bacterial arginine dihydrolase ArgZ suggests a "bond rotation" catalytic mechanism.
J.Biol.Chem., 295, 2020
|
|
4HTY
 
 | |
9BOR
 
 | | IkappaBzeta ankyrin repeat domain:NF-kappaB p50 homodimer complex at 2.0 Angstrom resolution | | Descriptor: | (2R,3S)-heptane-1,2,3-triol, NF-kappa-B inhibitor zeta, Nuclear factor NF-kappa-B p50 subunit | | Authors: | Rogers, W.E, Zhu, N, Huxford, T. | | Deposit date: | 2024-05-05 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural and biochemical analyses of the nuclear I kappa B zeta protein in complex with the NF-kappa B p50 homodimer.
Genes Dev., 38, 2024
|
|
8T7C
 
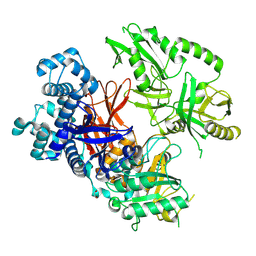 | | Crystal structure of human phospholipase C gamma 2 | | Descriptor: | 1,2-ETHANEDIOL, 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-2, CALCIUM ION | | Authors: | Chen, Y, Choi, H, Zhuang, N, Hu, L, Qian, D, Wang, J. | | Deposit date: | 2023-06-20 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The crystal and cryo-EM structures of PLCg2 reveal dynamic inter-domain recognitions in autoinhibition
To Be Published
|
|
8UPV
 
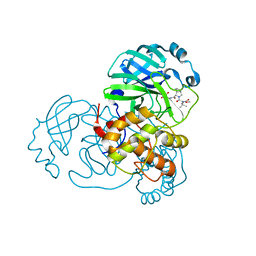 | | Structure of SARS-Cov2 3CLPro in complex with Compound 33 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6R)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
4IRB
 
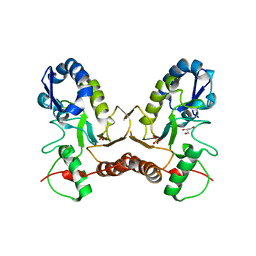 | | Crystal Structure of Vaccinia Virus Uracil DNA Glycosylase Mutant del171-172D4 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Schormann, N, Zhukovskaya, N, Sartmatova, D, Nuth, M, Ricciardi, R.P, Chattopadhyay, D. | | Deposit date: | 2013-01-14 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutations at the dimer interface affect both function and structure of the Vaccinia virus uracil DNA glycosylase
To be Published
|
|
8UPS
 
 | | Structure of SARS-Cov2 3CLPro in complex with Compound 5 | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-amino-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, PHOSPHATE ION | | Authors: | Wu, Y, Qiang, D, Zhuang, N, Krishnamurthy, H, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UPW
 
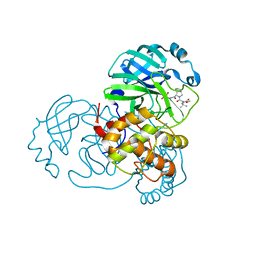 | | Structure of SARS-Cov2 3CLPro in complex with Compound 34 | | Descriptor: | 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S,6S)-6-fluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-23 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8UTE
 
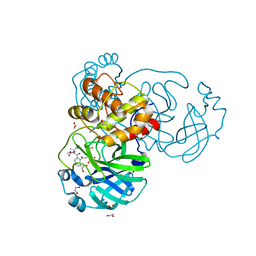 | | Structure of SARS-Cov2 3CLPro in complex with Compound 27 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, methyl {(2S)-1-[(1S,3aR,6aS)-1-{[(2R,3S)-6,6-difluoro-2-hydroxy-1-(methylamino)-1-oxoheptan-3-yl]carbamoyl}hexahydrocyclopenta[c]pyrrol-2(1H)-yl]-3,3-dimethyl-1-oxobutan-2-yl}carbamate | | Authors: | Krishnamurthy, H, Zhuang, N, Qiang, D, Wu, Y, Klein, D.J. | | Deposit date: | 2023-10-31 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Invention of MK-7845, a SARS-CoV-2 3CL Protease Inhibitor Employing a Novel Difluorinated Glutamine Mimic.
J.Med.Chem., 67, 2024
|
|
8TKL
 
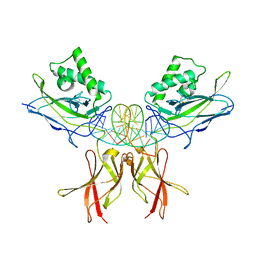 | | Murine NF-kappaB p50 Rel Homology Region homodimer in complex with a Test 16-mer kappaB-like DNA | | Descriptor: | Nuclear factor NF-kappa-B p50 subunit, Test 17-mer kappaB-like DNA | | Authors: | Mitchel, S, Mealka, M, Rogers, W.E, Milani, C, Acuna, L.M, Huxford, T. | | Deposit date: | 2023-07-25 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray Crystallographic Study of Preferred Spacing by the NF-kappa B p50 Homodimer on kappa B DNA.
Biomolecules, 13, 2023
|
|
5BXJ
 
 | | Complex of the Fk1 domain mutant A19T of FKBP51 with 4-Nitrophenol | | Descriptor: | P-NITROPHENOL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wu, D, Tao, X, Chen, Z, Han, J, Jia, W, Li, X, Wang, Z, He, Y.X. | | Deposit date: | 2015-06-09 | | Release date: | 2016-05-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The environmental endocrine disruptor p-nitrophenol interacts with FKBP51, a positive regulator of androgen receptor and inhibits androgen receptor signaling in human cells
J. Hazard. Mater., 307, 2016
|
|
3I9G
 
 | |
7FDK
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to mACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDH
 
 | | SARS-COV-2 Spike RBDMACSp25 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
7FDI
 
 | | SARS-COV-2 Spike RBDMACSp36 binding to hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Wang, X, Cao, L. | | Deposit date: | 2021-07-16 | | Release date: | 2021-08-25 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Characterization and structural basis of a lethal mouse-adapted SARS-CoV-2.
Nat Commun, 12, 2021
|
|
