5Z59
 
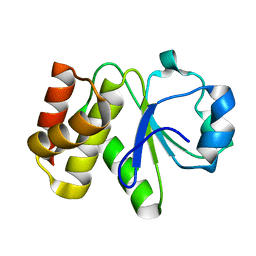 | | Crystal structure of Tk-PTP in the inactive form | | Descriptor: | Protein-tyrosine phosphatase | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
5Z5B
 
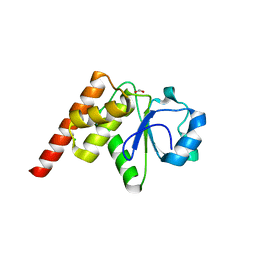 | | Crystal structure of Tk-PTP in the G95A mutant form | | Descriptor: | CHLORIDE ION, FORMIC ACID, Protein-tyrosine phosphatase | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-06-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
5Z5A
 
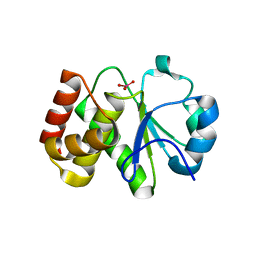 | | Crystal structure of Tk-PTP in the active form | | Descriptor: | Protein-tyrosine phosphatase, VANADATE ION | | Authors: | Ku, B, Yun, H.Y, Kim, S.J. | | Deposit date: | 2018-01-17 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural study reveals the temperature-dependent conformational flexibility of Tk-PTP, a protein tyrosine phosphatase from Thermococcus kodakaraensis KOD1
PLoS ONE, 13, 2018
|
|
7W91
 
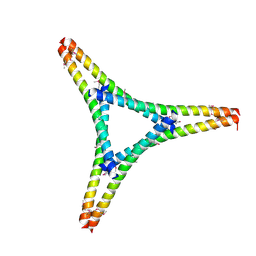 | | Residues 440-490 of centrosomal protein 63 | | Descriptor: | Centrosomal protein of 63 kDa | | Authors: | Yun, H.Y, Ku, B. | | Deposit date: | 2021-12-09 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.292 Å) | | Cite: | Architectural basis for cylindrical self-assembly governing Plk4-mediated centriole duplication in human cells.
Commun Biol, 6, 2023
|
|
6IWD
 
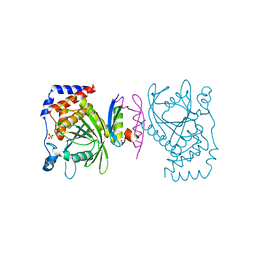 | | The PTP domain of human PTPN14 in a complex with the CR3 domain of HPV18 E7 | | Descriptor: | CHLORIDE ION, HPV18 E7, PHOSPHATE ION, ... | | Authors: | Yun, H.-Y, Kim, S.J, Ku, B. | | Deposit date: | 2018-12-05 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for recognition of the tumor suppressor protein PTPN14 by the oncoprotein E7 of human papillomavirus.
Plos Biol., 17, 2019
|
|
5XC5
 
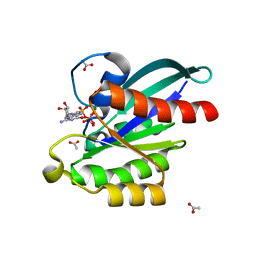 | | Crystal structure of Acanthamoeba polyphaga mimivirus Rab GTPase in complex with GTP | | Descriptor: | ACETATE ION, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ku, B, You, J.A, Kim, S.J. | | Deposit date: | 2017-03-22 | | Release date: | 2017-10-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Crystal structures of two forms of the Acanthamoeba polyphaga mimivirus Rab GTPase
Arch. Virol., 162, 2017
|
|
5XC3
 
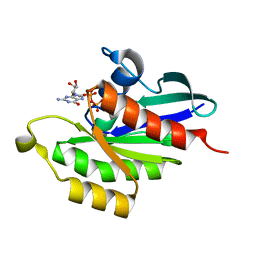 | |
7TUJ
 
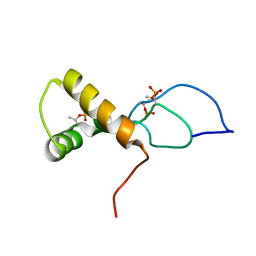 | | NMR solution structure of the phosphorylated MUS81-binding region from human SLX4 | | Descriptor: | Structure-specific endonuclease subunit SLX4 | | Authors: | Payliss, B.J, Reichheld, S.E, Lemak, A, Arrowsmith, C.H, Sharpe, S, Wyatt, H.D.M. | | Deposit date: | 2022-02-02 | | Release date: | 2022-10-19 | | Last modified: | 2022-11-09 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease.
Cell Rep, 41, 2022
|
|
7VZE
 
 | | Crystal structure of PTPN4 PDZ bound to the PBM of HPV16 E6 | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 4, the PDZ-binding motif of HPV16 E6 | | Authors: | Lee, H.S, Yun, H.-Y, Ku, B. | | Deposit date: | 2021-11-16 | | Release date: | 2022-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Structural and biochemical analysis of the PTPN4 PDZ domain bound to the C-terminal tail of the human papillomavirus E6 oncoprotein.
J.Microbiol, 60, 2022
|
|
7YTN
 
 | |
