1NBE
 
 | |
2QGF
 
 | | Structure of regulatory chain mutant H20A of asparate transcarbamoylase from E. coli | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Stec, B, Williams, M.K, Stieglitz, K.A, Kantrowitz, E.R. | | Deposit date: | 2007-06-28 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Comparison of two T-state structures of regulatory-chain mutants of Escherichia coli aspartate transcarbamoylase suggests that His20 and Asp19 modulate the response to heterotropic effectors.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2QG9
 
 | | Structure of a regulatory subunit mutant D19A of ATCase from E. coli | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, ZINC ION | | Authors: | Stec, B, Williams, M.K, Stieglitz, K.A, Kantrowitz, E.R. | | Deposit date: | 2007-06-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of two T-state structures of regulatory-chain mutants of Escherichia coli aspartate transcarbamoylase suggests that His20 and Asp19 modulate the response to heterotropic effectors.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1VZM
 
 | | OSTEOCALCIN FROM FISH ARGYROSOMUS REGIUS | | Descriptor: | MAGNESIUM ION, OSTEOCALCIN | | Authors: | Frazao, C, Simes, D.C, Coelho, R, Alves, D, Williamson, M.K, Price, P.A, Cancela, M.L, Carrondo, M.A. | | Deposit date: | 2004-05-21 | | Release date: | 2004-09-10 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Evidence of a Fourth Gla Residue in Fish Osteocalcin: Biological Implications
Biochemistry, 44, 2005
|
|
1R0C
 
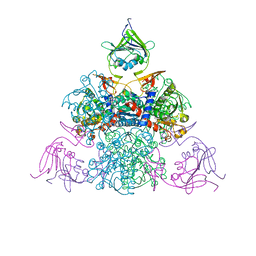 | |
7ZH2
 
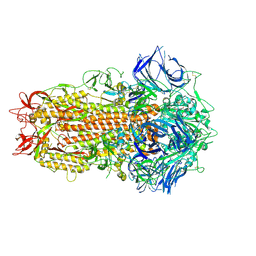 | | SARS CoV Spike protein, Closed C1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH5
 
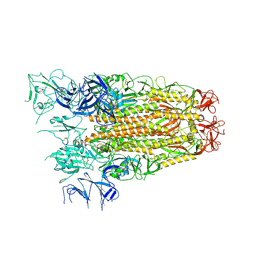 | | SARS CoV Spike protein, Open conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7ZH1
 
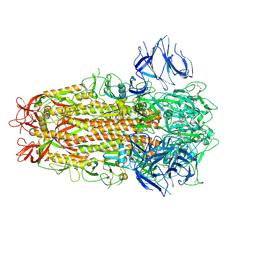 | | SARS CoV Spike protein, Closed C3 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
7OD3
 
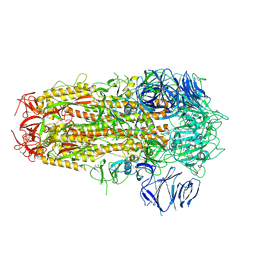 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-28 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7ODL
 
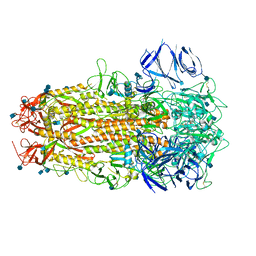 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-29 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
8C9N
 
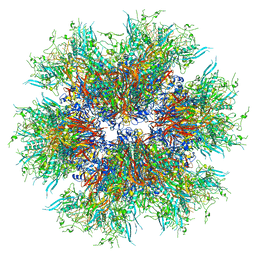 | | MiniCoV-ADDomer, a SARS-CoV-2 epitope presenting viral like particle | | Descriptor: | Penton protein | | Authors: | Bufton, J.C, Capin, J, Boruku, U, Garzoni, F, Schaffitzel, C, Berger, I. | | Deposit date: | 2023-01-23 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | In vitro generated antibodies guide thermostable ADDomer nanoparticle design for nasal vaccination and passive immunization against SARS-CoV-2.
Antib Ther, 6, 2023
|
|
7JJC
 
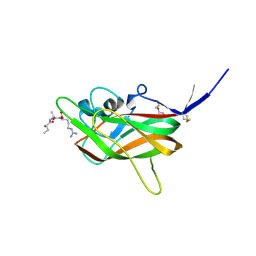 | |
