8R6K
 
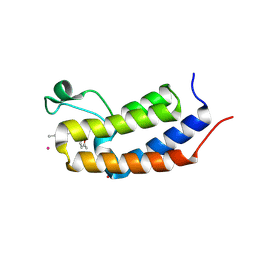 | | Crystal structure of Candida glabrata Bdf1 bromodomain 1 bound to a phenyltriazine ligand | | Descriptor: | 6-methyl-~{N}-[(5-methylfuran-2-yl)methyl]-3-(4-methylphenyl)-1,2,4-triazin-5-amine, Candida glabrata strain CBS138 chromosome C complete sequence, PHOSPHATE ION, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6I
 
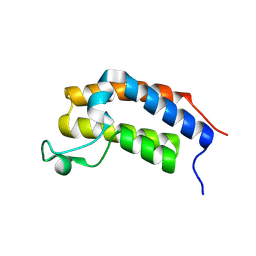 | |
8R6J
 
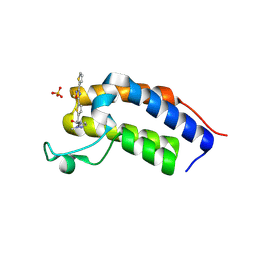 | | Crystal structure of Candida glabrata Bdf1 bromodomain 1 bound to a pyrazole ligand | | Descriptor: | 2-methyl-~{N}-[[5-(3-thiophen-2-yl-1,2,4-oxadiazol-5-yl)thiophen-2-yl]methyl]pyrazole-3-carboxamide, Candida glabrata strain CBS138 chromosome C complete sequence, SULFATE ION | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6M
 
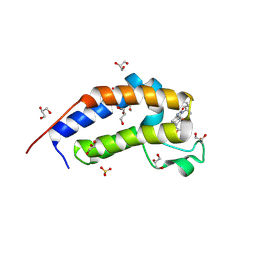 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 bound to I-BET151 | | Descriptor: | 7-(3,5-DIMETHYL-1,2-OXAZOL-4-YL)-8-METHOXY-1-[(1R)-1-(PYRIDIN-2-YL)ETHYL]-1H,2H,3H-IMIDAZO[4,5-C]QUINOLIN-2-ONE, Candida glabrata strain CBS138 chromosome C complete sequence, GLYCEROL, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6N
 
 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 bound to a pyridoindole ligand | | Descriptor: | 2-ethanoyl-~{N}-(4-morpholin-4-ylphenyl)-1,3,4,5-tetrahydropyrido[4,3-b]indole-8-carboxamide, Candida glabrata strain CBS138 chromosome C complete sequence | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
8R6L
 
 | | Crystal structure of Candida glabrata Bdf1 bromodomain 2 in the unbound state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Candida glabrata strain CBS138 chromosome C complete sequence, ... | | Authors: | Petosa, C, Wei, K, McKenna, C.E, Govin, J. | | Deposit date: | 2023-11-22 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Humanized Candida and NanoBiT Assays Expedite Discovery of Bdf1 Bromodomain Inhibitors with Antifungal Potential against invasive Candida infection
Advanced Science, 2024
|
|
6YEP
 
 | |
6YCV
 
 | |
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
7PER
 
 | | Model of the inner ring of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup155, Nuclear pore complex protein Nup205, Nuclear pore complex protein Nup93, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
3GFP
 
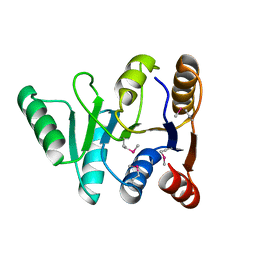 | | Structure of the C-terminal domain of the DEAD-box protein Dbp5 | | Descriptor: | DEAD box protein 5 | | Authors: | Erzberger, J.P, Dossani, Z.Y, Weirich, C.S, Weis, K, Berger, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
1XIP
 
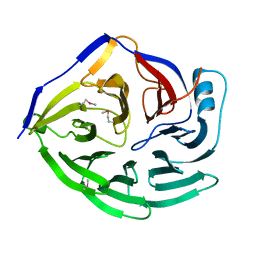 | |
1QGR
 
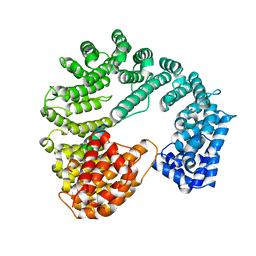 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA (II CRYSTAL FORM, GROWN AT LOW PH) | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-05-04 | | Release date: | 1999-05-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
1QGK
 
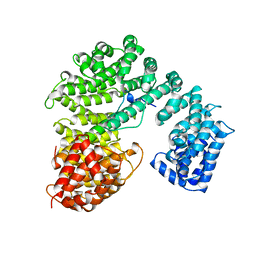 | | STRUCTURE OF IMPORTIN BETA BOUND TO THE IBB DOMAIN OF IMPORTIN ALPHA | | Descriptor: | PROTEIN (IMPORTIN ALPHA-2 SUBUNIT), PROTEIN (IMPORTIN BETA SUBUNIT) | | Authors: | Cingolani, G, Petosa, C, Weis, K, Muller, C.W. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of importin-beta bound to the IBB domain of importin-alpha.
Nature, 399, 1999
|
|
3PEU
 
 | | S. cerevisiae Dbp5 L327V C-terminal domain bound to Gle1 H337R and IP6 | | Descriptor: | ATP-dependent RNA helicase DBP5, GLYCEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2020-10-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEW
 
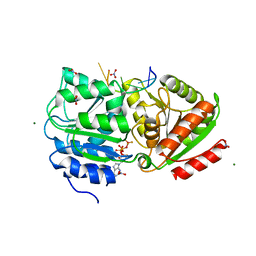 | | S. cerevisiae Dbp5 L327V bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEV
 
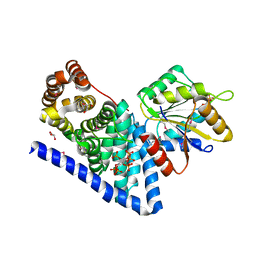 | | S. cerevisiae Dbp5 L327V C-terminal domain bound to Gle1 and IP6 | | Descriptor: | ATP-dependent RNA helicase DBP5, GLYCEROL, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3PEY
 
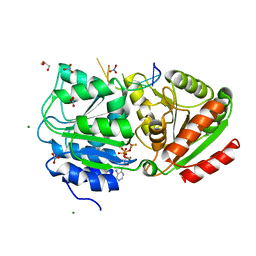 | | S. cerevisiae Dbp5 bound to RNA and ADP BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2010-10-27 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP(6) in mRNA export.
Nature, 472, 2011
|
|
3RRN
 
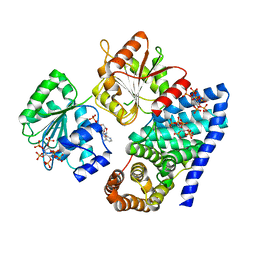 | | S. cerevisiae dbp5 l327v bound to gle1 h337r and ip6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.001 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
3RRM
 
 | | S. cerevisiae dbp5 l327v bound to nup159, gle1 h337r, ip6 and adp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DBP5, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Montpetit, B, Thomsen, N.D, Helmke, K.J, Seeliger, M.A, Berger, J.M, Weis, K. | | Deposit date: | 2011-04-29 | | Release date: | 2011-05-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export.
Nature, 472, 2011
|
|
1GS9
 
 | | Apolipoprotein E4, 22k domain | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Verderame, J.R, Kantardjieff, K, Segelke, B, Weisgraber, K, Rupp, B. | | Deposit date: | 2002-01-02 | | Release date: | 2003-06-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the 22K Domain of Human Apolipoprotein E4
To be Published
|
|
6ZL9
 
 | |
6ERL
 
 | |
6R9K
 
 | |
6R9L
 
 | |
