6T9J
 
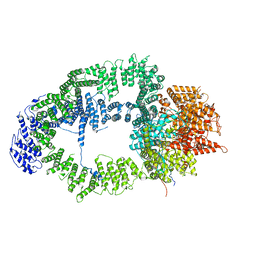 | | SAGA Tra1 module | | Descriptor: | Transcription factor SPT20, Transcription initiation factor TFIID subunit 12, Transcription-associated protein 1 | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6T9I
 
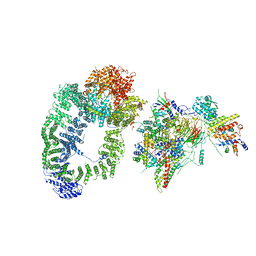 | | cryo-EM structure of transcription coactivator SAGA | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
6T9L
 
 | |
8B38
 
 | |
8B3J
 
 | |
8B59
 
 | |
8B4Z
 
 | |
2YW8
 
 | | Crystal structure of human RUN and FYVE domain-containing protein | | Descriptor: | RUN and FYVE domain-containing protein 1, SULFATE ION, ZINC ION | | Authors: | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2008-04-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human RUN and FYVE domain-containing protein
To be Published
|
|
2YV6
 
 | | Crystal structure of human Bcl-2 family protein Bak | | Descriptor: | Bcl-2 homologous antagonist/killer, SULFATE ION | | Authors: | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-09 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel dimerization mode of the human Bcl-2 family protein Bak, a mitochondrial apoptosis regulator.
J.Struct.Biol., 166, 2009
|
|
6HBA
 
 | | Crystal Structure of the small subunit-like domain 1 of CcmM from Synechococcus elongatus (strain PCC 7942), thiol-oxidized form | | Descriptor: | Carbon dioxide concentrating mechanism protein CcmM | | Authors: | Wang, H, Yan, X, Aigner, H, Bracher, A, Nguyen, N.D, Hee, W.Y, Long, B.M, Price, G.D, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-08-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rubisco condensate formation by CcmM in beta-carboxysome biogenesis.
Nature, 566, 2019
|
|
7OHA
 
 | | nucleosome with TBP and TFIIA bound at SHL +2 | | Descriptor: | DNA (122-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OH9
 
 | | Nucleosome with TBP and TFIIA bound at SHL -6 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHB
 
 | | TBP-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-10 | | Release date: | 2021-07-28 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHC
 
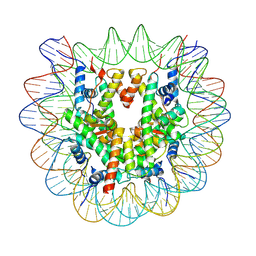 | |
7OS7
 
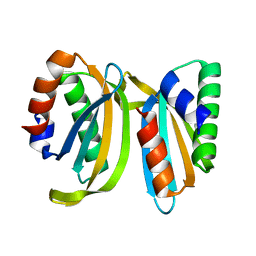 | | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant | | Descriptor: | 30S ribosomal protein S6,30S ribosomal protein S6 | | Authors: | Wang, H, Logan, D.T, Oliveberg, M. | | Deposit date: | 2021-06-08 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Circular permutant of ribosomal protein S6, swap helix 2, L75A, A92K mutant
To Be Published
|
|
6T9K
 
 | | SAGA Core module | | Descriptor: | Protein SPT3, SAGA-associated factor 73, Transcription factor SPT20, ... | | Authors: | Wang, H, Cheung, A, Cramer, P. | | Deposit date: | 2019-10-28 | | Release date: | 2020-01-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the transcription coactivator SAGA.
Nature, 577, 2020
|
|
2YV5
 
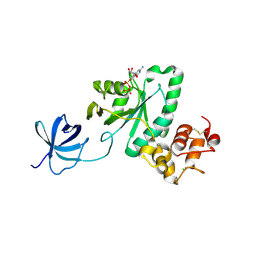 | | Crystal structure of Yjeq from Aquifex aeolicus | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, YjeQ protein, ... | | Authors: | Wang, H, Kaminishi, T, Hanawa-Suetsugu, K, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-09 | | Release date: | 2008-04-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of YjeQ from Aquifex aeolicus
To be Published
|
|
2YVR
 
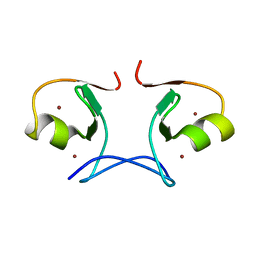 | | Crystal structure of MS1043 | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Wang, H, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-13 | | Release date: | 2008-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of MS1043
To be Published
|
|
2Z0I
 
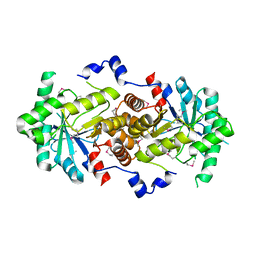 | | Crystal Structure of 5-aminolevulinic acid dehydratase (ALAD) from Mus musculus | | Descriptor: | Delta-aminolevulinic acid dehydratase | | Authors: | Wang, H, Xie, Y, Kawazoe, M, Kishishita, S, Murayama, K, Takemoto, C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-07 | | Release date: | 2008-05-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of 5-aminolevulinic acid dehydratase (ALAD) from Mus musculus
To be Published
|
|
2CSL
 
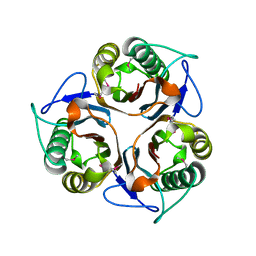 | | Crystal structure of TTHA0137 from Thermus Thermophilus HB8 | | Descriptor: | protein translation initiation inhibitor | | Authors: | Wang, H, Murayama, K, Terada, T, Chen, L, Jin, Z, Chrzas, J, Liu, Z.J, Wang, B.C, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-22 | | Release date: | 2005-11-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of TTHA0137 from Thermus Thermophilus HB8
To be Published
|
|
3ECN
 
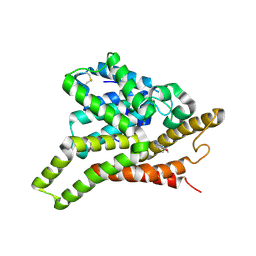 | | Crystal structure of PDE8A catalytic domain in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
3ECM
 
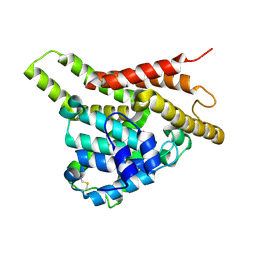 | | Crystal structure of the unliganded PDE8A catalytic domain | | Descriptor: | High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
3NGN
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CCR4-NOT transcription complex subunit 6-like | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGO
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with poly(A) DNA | | Descriptor: | 5'-D(*AP*AP*AP*A)-3', CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3NGQ
 
 | | Crystal structure of the human CNOT6L nuclease domain | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
