5L0Y
 
 | |
5L0W
 
 | |
3FHN
 
 | | Structure of Tip20p | | Descriptor: | Protein transport protein TIP20 | | Authors: | Tripathi, A, Ren, Y, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2008-12-09 | | Release date: | 2009-01-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex.
Nat.Struct.Mol.Biol., 16, 2009
|
|
1YUC
 
 | | Human Nuclear Receptor Liver Receptor Homologue-1, LRH-1, Bound to Phospholipid and a Fragment of Human SHP | | Descriptor: | GLYCEROL, L-ALPHA-PHOSPHATIDYL-BETA-OLEOYL-GAMMA-PALMITOYL-PHOSPHATIDYLETHANOLAMINE, Nuclear receptor 0B2, ... | | Authors: | Ortlund, E.A, Yoonkwang, L, Solomon, I.H, Hager, J.M, Safi, R, Choi, Y, Guan, Z, Tripathy, A, Raetz, C.R.H, McDonnell, D.P, Moore, D.D, Redinbo, M.R. | | Deposit date: | 2005-02-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP
Nat.Struct.Mol.Biol., 12, 2005
|
|
3ZGZ
 
 | | Ternary complex of E. coli leucyl-tRNA synthetase, tRNA(leu) and toxic moiety from agrocin 84 (TM84) in aminoacylation-like conformation | | Descriptor: | LEUCINE--TRNA LIGASE, MAGNESIUM ION, TRNA-LEU UAA ISOACCEPTOR, ... | | Authors: | Chopra, S, Palencia, A, Virus, C, Tripathy, A, Temple, B.R, Velazquez-Campoy, A, Cusack, S, Reader, J.S. | | Deposit date: | 2012-12-19 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Plant Tumour Biocontrol Agent Employs a tRNA-Dependent Mechanism to Inhibit Leucyl-tRNA Synthetase
Nat.Commun., 4, 2013
|
|
3ZY7
 
 | | Crystal structure of computationally redesigned gamma-adaptin appendage domain forming a symmetric homodimer | | Descriptor: | AP-1 COMPLEX SUBUNIT GAMMA-1, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL | | Authors: | Stranges, P.B, Machius, M, Miley, M.J, Tripathy, A, Kuhlman, B. | | Deposit date: | 2011-08-17 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Computational Design of a Symmetric Homodimer Using Beta-Strand Assembly.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6ECA
 
 | | Lactobacillus rhamnosus Beta-glucuronidase | | Descriptor: | Beta-glucuronidase, CHLORIDE ION, GLYCEROL | | Authors: | Biernat, K.A, Pellock, S.J, Bhatt, A.P, Bivins, M.M, Walton, W.G, Tran, B.N.T, Wei, L, Snider, M.C, Cesmat, A.P, Tripathy, A, Erie, D.A, Redinbo, M.R.R. | | Deposit date: | 2018-08-07 | | Release date: | 2019-02-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.853 Å) | | Cite: | Structure, function, and inhibition of drug reactivating human gut microbial beta-glucuronidases.
Sci Rep, 9, 2019
|
|
6DII
 
 | | Structure of Arabidopsis Fatty Acid Amide Hydrolase in Complex with methyl linolenyl fluorophosphonate | | Descriptor: | Fatty acid amide hydrolase, methyl-9Z,12Z,15Z-octadecatrienylphosphonofluoridate | | Authors: | Aziz, M, Wang, X, Tripathi, A, Bankaitis, V, Chapman, K.D. | | Deposit date: | 2018-05-23 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides.
J.Biol.Chem., 294, 2019
|
|
6DHV
 
 | | Structure of Arabidopsis Fatty Acid Amide Hydrolase | | Descriptor: | Fatty acid amide hydrolase | | Authors: | Aziz, M, Wang, X, Tripathi, A, Bankaitis, V, Chapman, K.D. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides.
J.Biol.Chem., 294, 2019
|
|
8GKV
 
 | | Crystal structure of anti-adaptor IraP that regulates RpoS proteolysis | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Shaw, G.X, Gan, J, Suburaman, P, Battesti, A, Zhou, Y.N, Wickner, S, Gottesman, S, Ji, X. | | Deposit date: | 2023-03-20 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural and functional study of anti-adaptor IraP-mediated regulation of RpoS proteolysis
to be published
|
|
5WM7
 
 | | Crystal Structure of CahJ in Complex with AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.777 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5WM6
 
 | | Crystal Structure of CahJ in Complex with Benzoyl Adenylate | | Descriptor: | 5'-O-[(R)-(benzoyloxy)(hydroxy)phosphoryl]adenosine, ACETATE ION, MAGNESIUM ION, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5WM2
 
 | |
5WM4
 
 | | Crystal Structure of CahJ in Complex with 6-Methylsalicyl Adenylate | | Descriptor: | 9-(5-O-{(S)-hydroxy[(2-hydroxy-6-methylbenzene-1-carbonyl)oxy]phosphoryl}-alpha-L-lyxofuranosyl)-9H-purin-6-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5WM5
 
 | | Crystal Structure of CahJ in Complex with 5-Methylsalicyl Adenylate | | Descriptor: | 9-(5-O-{(S)-hydroxy[(2-hydroxy-5-methylbenzene-1-carbonyl)oxy]phosphoryl}-alpha-L-lyxofuranosyl)-9H-purin-6-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
5WM3
 
 | | Crystal Structure of CahJ in Complex with Salicyl Adenylate | | Descriptor: | 9-(5-O-{(S)-hydroxy[(2-hydroxybenzene-1-carbonyl)oxy]phosphoryl}-alpha-L-lyxofuranosyl)-9H-purin-6-amine, ACETATE ION, GLYCEROL, ... | | Authors: | Sikkema, A.P, Smith, J.L. | | Deposit date: | 2017-07-28 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | A Defined and Flexible Pocket Explains Aryl Substrate Promiscuity of the Cahuitamycin Starter Unit-Activating Enzyme CahJ.
Chembiochem, 19, 2018
|
|
3ETU
 
 | | Crystal structure of yeast Dsl1p | | Descriptor: | Protein transport protein DSL1 | | Authors: | Ren, Y, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2008-10-08 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3ETV
 
 | | Crystal structure of a Tip20p-Dsl1p fusion protein | | Descriptor: | Protein transport protein TIP20, Protein transport protein DSL1 chimera | | Authors: | Ren, Y, Jeffrey, P.D, Hughson, F.M. | | Deposit date: | 2008-10-08 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural characterization of Tip20p and Dsl1p, subunits of the Dsl1p vesicle tethering complex.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3RJW
 
 | | Crystal structure of histone lysine methyltransferase g9a with an inhibitor | | Descriptor: | 2-cyclohexyl-6-methoxy-N-[1-(1-methylethyl)piperidin-4-yl]-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, Histone-lysine N-methyltransferase EHMT2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Dong, A, Wasney, G.A, Tempel, W, Liu, F, Barsyte, D, Allali-Hassani, A, Chen, X, Chau, I, Hajian, T, Senisterra, G, Chavda, N, Arora, K, Siarheyeva, A, Kireev, D.B, Herold, J.M, Bochkarev, A, Bountra, C, Weigelt, J, Edwards, A.M, Frye, S.V, Arrowsmith, C.H, Brown, P.J, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-04-15 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells.
Nat.Chem.Biol., 7, 2011
|
|
6D50
 
 | | Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, CALCIUM ION, Glycosyl hydrolases family 2, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D89
 
 | |
6D7J
 
 | |
6D7F
 
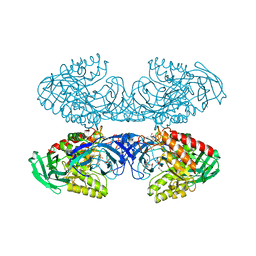 | | Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide | | Descriptor: | Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D8K
 
 | |
4HT6
 
 | |
