6Z2Z
 
 | |
6Z2U
 
 | |
8AGI
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
8AGJ
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112A in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
8AGL
 
 | | Structure of human Heat shock protein 90-alpha N-terminal domain (Hsp90-NTD) variant K112R in complex with JMC31 | | Descriptor: | 1-[2,4-bis(oxidanyl)-5-propan-2-yl-phenyl]-5-[4-[(cyclohexylmethylamino)methyl]phenyl]-~{N}-ethyl-1,2,3-triazole-4-carboxamide, Heat shock protein HSP 90-alpha | | Authors: | Tassone, G, Pozzi, C, Mazzorana, M, Mangani, S, Maramai, S. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Human Heat Shock Protein 90 N-Terminal Domain and Its Variants K112R and K112A in Complex with a Potent 1,2,3-Triazole-Based Inhibitor.
Int J Mol Sci, 23, 2022
|
|
9ETC
 
 | |
9ETG
 
 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with CA-M11 | | Descriptor: | Fatty acid-binding protein, liver, [4-[(2-azanyl-4-oxidanylidene-1,3-thiazol-5-yl)methyl]phenyl] (4~{R})-4-[(3~{R},5~{R},7~{R},8~{R},9~{S},10~{S},12~{S},13~{R},14~{S},17~{R})-10,13-dimethyl-3,7,12-tris(oxidanyl)-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1~{H}-cyclopenta[a]phenanthren-17-yl]pentanoate | | Authors: | Tassone, G, Pozzi, C, Maramai, S. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
9FXG
 
 | | Crystal structure of double mutant Y115E Y117E human Glutaminyl Cyclase in apo-state | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective.
Int J Mol Sci, 25, 2024
|
|
9FXI
 
 | | Crystal structure of cobalt(II)-substituted double mutant Y115E Y117E human Glutaminyl Cyclase in complex with SEN177 | | Descriptor: | 2-fluoranyl-5-[2-[4-(4-methyl-1,2,4-triazol-3-yl)piperidin-1-yl]pyridin-3-yl]pyridine, COBALT (II) ION, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective.
Int J Mol Sci, 25, 2024
|
|
9FXJ
 
 | | Crystal structure of cobalt(II)-substituted double mutant Y115E Y117E human Glutaminyl Cyclase in complex with PBD-150 | | Descriptor: | 1-(3,4-dimethoxyphenyl)-3-[3-(1H-imidazol-1-yl)propyl]thiourea, COBALT (II) ION, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective.
Int J Mol Sci, 25, 2024
|
|
9FXH
 
 | | Crystal structure of cobalt(II)-substituted double mutant Y115E Y117E human Glutaminyl Cyclase | | Descriptor: | COBALT (II) ION, GLYCEROL, Glutaminyl-peptide cyclotransferase, ... | | Authors: | Tassone, G, Pozzi, C, Mangani, S. | | Deposit date: | 2024-07-01 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Metal Ion Binding to Human Glutaminyl Cyclase: A Structural Perspective.
Int J Mol Sci, 25, 2024
|
|
9ETD
 
 | |
9ETE
 
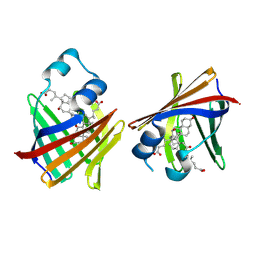 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, Fatty acid-binding protein, liver | | Authors: | Tassone, G, Pozzi, C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
9ETF
 
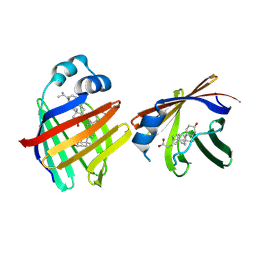 | | Crystal structure of recombinant chicken liver Bile Acid Binding Protein (cL-BABP) in complex with lithocholic acid | | Descriptor: | (3beta,5beta,14beta,17alpha)-3-hydroxycholan-24-oic acid, Fatty acid-binding protein, liver | | Authors: | Tassone, G, Pozzi, C. | | Deposit date: | 2024-03-26 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploiting the bile acid binding protein as transporter of a Cholic Acid/Mirin bioconjugate for potential applications in liver cancer therapy.
Sci Rep, 14, 2024
|
|
6ZSW
 
 | |
6ZSX
 
 | |
7OPJ
 
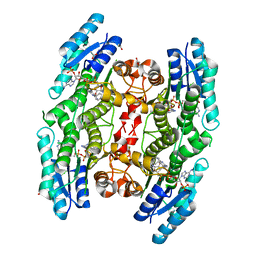 | | Trypanosoma brucei PTR1 (TbPTR1) in complex with pyrimethamine | | Descriptor: | 5-(4-CHLORO-PHENYL)-6-ETHYL-PYRIMIDINE-2,4-DIAMINE, ACETATE ION, GLYCEROL, ... | | Authors: | Tassone, G, Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2021-05-31 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Evidence of Pyrimethamine and Cycloguanil Analogues as Dual Inhibitors of Trypanosoma brucei Pteridine Reductase and Dihydrofolate Reductase.
Pharmaceuticals, 14, 2021
|
|
6ZRJ
 
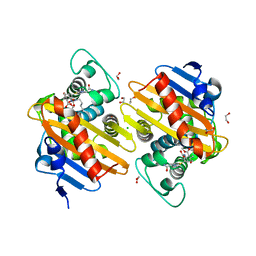 | | Crystal structure of class D Beta-lactamase OXA-48 in complex with ertapenem | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Docquier, J.D, Mangani, S. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRG
 
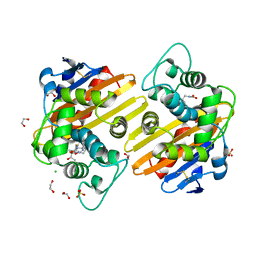 | | Crystal structure of OXA-10loop48 in complex with hydrolyzed doripenem | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-({[amino(dihydroxy)methyl]amino}methyl)pyrrolidin-3-yl]sulfanyl}-5-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-methyl-4,5-dihydro-1H-pyrrole-2-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Docquier, J.D, Mangani, S. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRH
 
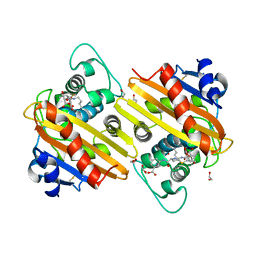 | | Crystal structure of OXA-10loop24 in complex with ertapenem | | Descriptor: | (2S,3R,4S)-4-({(3S,5S)-5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl}sulfanyl)-2-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRI
 
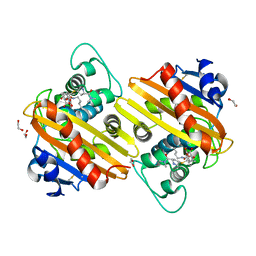 | | Crystal structure of OXA-10loop24 in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZRP
 
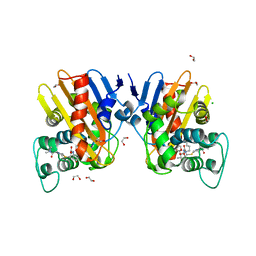 | | Crystal structure of class D Beta-lactamase OXA-48 in complex with meropenem | | Descriptor: | (2S,3R,4S)-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-2-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-3-methyl-3,4-dihydro-2H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, De Luca, F, Pozzi, C, Di Pisa, F, Benvenuti, M, Mangani, S, Doquier, J.D. | | Deposit date: | 2020-07-14 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
6ZW2
 
 | | Crystal structure of OXA-10loop48 in complex with hydrolyzed meropenem | | Descriptor: | (2S,3R)-2-[(2S,3R)-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfan yl-3-methyl-2,3-dihydro-1H-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Beta-lactamase, ... | | Authors: | Tassone, G, Di Pisa, F, Benvenuti, M, De Luca, F, Pozzi, C, Mangani, S, Docquier, J.D. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanistic insights into carbapenem hydrolysis by OXA-48 and the OXA10-derived hybrids OXA-10 loop24 and loop48
To Be Published
|
|
7O0K
 
 | |
7O0J
 
 | |
